Abstract
BACKGROUND: Determination of isocitrate dehydrogenase (IDH) status and, if IDH-mutant, assessing 1p19q codeletion are an important component of diagnosis of World Health Organization grades II/III or lower-grade gliomas. This has led to research into noninvasively correlating imaging features (“radiomics”) with genetic status.
PURPOSE: Our aim was to perform a diagnostic test accuracy systematic review for classifying IDH and 1p19q status using MR imaging radiomics, to provide future directions for integration into clinical radiology.
DATA SOURCES: Ovid (MEDLINE), Scopus, and the Web of Science were searched in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses for Diagnostic Test Accuracy guidelines.
STUDY SELECTION: Fourteen journal articles were selected that included 1655 lower-grade gliomas classified by their IDH and/or 1p19q status from MR imaging radiomic features.
DATA ANALYSIS: For each article, the classification of IDH and/or 1p19q status using MR imaging radiomics was evaluated using the area under curve or descriptive statistics. Quality assessment was performed with the Quality Assessment of Diagnostic Accuracy Studies 2 tool and the radiomics quality score.
DATA SYNTHESIS: The best classifier of IDH status was with conventional radiomics in combination with convolutional neural network–derived features (area under the curve = 0.95, 94.4% sensitivity, 86.7% specificity). Optimal classification of 1p19q status occurred with texture-based radiomics (area under the curve = 0.96, 90% sensitivity, 89% specificity).
LIMITATIONS: A meta-analysis showed high heterogeneity due to the uniqueness of radiomic pipelines.
CONCLUSIONS: Radiogenomics is a potential alternative to standard invasive biopsy techniques for determination of IDH and 1p19q status in lower-grade gliomas but requires translational research for clinical uptake.
ABBREVIATIONS:
- AI
- artificial intelligence
- AUC
- area under the curve
- CNN
- convolutional neural network
- IDH
- isocitrate dehydrogenase
- IDH-mut
- IDH-mutant
- LGG
- lower-grade gliomas
- ML
- machine learning
- PRISMA-DTA
- Preferred Reporting Items for Systematic Reviews and Meta-Analyses for Diagnostic Test Accuracy
- QUADAS-2
- Quality Assessment of Diagnostic Accuracy Studies 2
- RQS
- radiomics quality score
- SVM
- support vector machine
- VASARI
- Visually Accessible Rembrandt Images
- WHO
- World Health Organization
Lower-grade gliomas (LGG), World Health Organization (WHO) grades II/III, are diffusely infiltrative tumors of the CNS. With time, these tumors typically progress to glioblastoma (WHO grade IV), which has a median survival of only 12–18 months despite treatment.1 A growing understanding of the prognostic and therapeutic importance of molecular markers has led to their incorporation into the 2016 WHO classification, and they now constitute a key component of the diagnosis of LGG.2 The 2 key markers of LGG are isocitrate dehydrogenase (IDH), with tumors classified as either IDH-mutant (IDH-mut) or IDH-wild-type, and 1p19q, with 1p19q-codeletion representing a combined loss of both the short arm of chromosome 1 and the long arm of chromosome 19.
Determining IDH and 1p19q status is invasive, requiring a tissue specimen via stereotactic biopsy or definitive resection, with the associated operative risks3 and possibility of sampling error. While the possibility of sampling error is perhaps of greatest relevance to the determination of tumor grade,4 it is also relevant to the determination of tumor genetic status.5,6 For example, IDH sequencing may be falsely negative if there are few glioma cells within the sample,5 and intratumoral genetic heterogeneity can occur.6 These considerations have led to research into characterizing IDH and 1p19q status by imaging, known as “radiogenomics” or “imaging genomics.” The most specific visual MR imaging feature is the “T2-FLAIR mismatch sign,” which has been shown to predict an IDH-mut 1p19q-codeletion gliomas with 100% specificity and high interobserver correlation (κ = 0.38–0.88).7⇓-9 Other useful features include the presence of calcification (suggestive of a 1p19q-codeletion glioma)8,9 and homogeneous signal (likely 1p19q-intact).10 While some features such as >50% T2-FLAIR mismatch and the presence of calcification have high interobserver correlation, other features are limited by greater variability in interpretation. Furthermore, a substantial proportion (29%–37%) of gliomas do not exhibit these features, limiting sensitivity.8
Artificial intelligence (AI) is emerging as a solution to the limitations of conventional visual assessment. AI techniques may identify features hidden to the naked eye by extracting data from images and relating them to outcomes. Given the inherent signal and volume heterogeneity of gliomas, a perceived signature or pattern may be modelled to genetic, clinical, and biochemical outcomes.11 Features can be learned from the image or predefined. The field of radiomics involves the extraction of predefined features such as shape, intensity, and texture from a segmented (tumor) volume of interest.12 This is opposed to deep learning–derived features, which are identified without human predefinition. Radiomic features can be correlated with genetic status through a subset of AI known as machine learning (ML). The ML algorithm is trained to a clinical outcome via a training dataset and validated using a testing/validation dataset. Extracted radiomic features undergo selection and can then be related to molecular markers such as IDH and 1p19q, providing a more objective method of radiogenomic correlation.
Radiomic analysis has several advantages compared with human observers, including the ability to rapidly assess multiple imaging features, less interobserver variability,13 and potentially higher sensitivity and specificity. The aim of this article was to perform a systemic review of the use of MR imaging radiomics for the classification of IDH and 1p19q status in LGG.
MATERIALS AND METHODS
Search methodology and study synthesis were performed in line with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses for Diagnostic Test Accuracy (PRISMA-DTA) checklist.14 The search was performed on the Web of Science, Ovid (MEDLINE), and Scopus on April 18, 2020. Online Table 1 summarizes the search strategy. Search terms were developed from the PICO framework and Medical Subject Headings, which included terms relating to radiomics or radiogenomics, gliomas, and IDH/1p19q status. The PRISMA flowchart is available in Online Fig 1.
Study Selection
Studies were included if they were original research articles relating radiomic features to IDH and/or 1p19q status in LGG (WHO grades II/III) with pathologic confirmation. Studies were excluded under the following circumstances: 1) They investigated the effects of radiogenomic pipelines on factors that affect imaging quality rather than assessing diagnostic potential or 2) they included imaging modalities other than MR imaging because recent literature has not shown superior outcomes.15 There was no restriction on study date.
The references were imported from the Web of Science, Ovid (MEDLINE), and Scopus into EndNote (Version X9; https://www.endnote.com/product-details/). Duplicates were removed using the “Find Duplicates” function in EndNote and manual review of the reference list. Two independent authors (A.P.B. and J.K.) screened the titles and abstracts for eligibility. The full texts were then screened. When questions arose regarding inclusion of articles, these were resolved through discussion between both authors responsible for data extraction (experience: A.P.B., medical doctor with a master’s degree in medical imaging analysis, and J.K., medical doctor with 4 years’ clinical experience). Ties were to be reviewed together with the senior author, but none were encountered.
Data Collection and Analysis
The primary outcome was the classification of IDH and/or 1p19q status by MR imaging radiomics. This was based on the receiver operating or precision recall curve and associated sensitivity (%), specificity (%), and area under the curve (AUC) if available. The AUC is presented as a value between 0.5 and 1, with 1 representing perfect classification (and 100% sensitivity and specificity). For studies that did not include ML in the pipeline, descriptive statistics (for example, mean and SD with t testing) were also included. Only significant findings for descriptive statistics were reported or the highest AUC for ML classifiers, given that some studies related numerous radiomic features to genetic status (IDH and/or 1p19q) or reported a considerable number of ML classifiers. If training and validation set data were reported, only the validation set was used. Secondary outcome measures were related to pipeline features and included the number of lesions, imaging sequences and segmentation method, features and their selection method, ML classifier, genetic status, and WHO tumor grade. A meta-analysis using random effects16 was performed on AUC values with 95% confidence intervals when available in MedCalc (MedCalc Software). A Higgins I2 index of heterogeneity was reported, in which 0% represents no heterogeneity and 100% represents maximum heterogeneity.
Quality Assessment
Quality assessment was performed using the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) tool and the radiomics quality score (RQS).12 The QUADAS-2 scoring system was developed to assess bias and the applicability of diagnostic-accuracy studies.17 The RQS is specific to radiomics and is based on the Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis initiative, which examines domains of application for predictive models.18 Application of RQS and QUADAS-2 was performed by discussion between A.P.B. and J.K. A κ statistic19 was considered for the RQS, similar to that used in previous studies;20 however, for quantitatively-defined criteria, it was determined that resolution by discussion would be superior.21
RESULTS
The initial search obtained 610 articles; 431 articles were from Ovid (MEDLINE); 111, from Scopus; and 68, from the Web of Science. After duplicates were removed, a total of 532 articles remained. The articles were screened by title and abstract, and 18 remained. Full texts were reviewed, and 14 articles22-35 fit the review question and inclusion criteria. The publication dates of the 14 included studies22⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓-35 ranged from 2017 to 2020. A total of 1655 LGG were analyzed. Online Table 2 summarizes the pipeline features for each study.
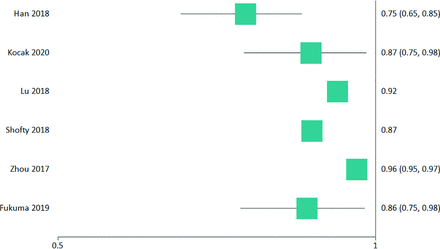
All segmentations incorporated manual components except for 2 studies, both of which used convolutional neural network (CNN)-based segmentation.28,32 Standard imaging sequences included pre- and postcontrast T1WI, T2WI, and FLAIR. ADC,23,25,30 cerebral blood flow/volume,28,30 DTI,29 and exponential ADC30 were used as adjuncts in some studies. Radiomic features were extracted most commonly by programs developed in-house on the Matlab software platform (MathWorks).23,25,27 AlexNet (https://www.mygreatlearning.com/blog/alexnet-the-first-cnn-to-win-image-net/) was used in 1 study for deep learning–derived features in the highest discriminating pipeline.27 The most common method of feature selection was support vector machine (SVM)–recursive feature elimination,25,30,33,34 followed by a Student t test.27-29 All categories of radiomic features were used. Two studies did not use ML.23,24 Most studies assessed WHO grade II and III LGG,22⇓⇓⇓⇓⇓⇓⇓⇓-30,32⇓-34 apart from one that assessed only WHO grade II LGG.31 Table 1 demonstrates the derived aims and key findings of studies that examined the IDH status of LGG, while Table 2 summarizes studies examining the 1p19q status of IDH-mut LGG. Figures 1 and 2 provide the associated forest plots for studies assessing IDH and 1p19q, respectively. Further details are provided in the online material. A meta-analysis on IDH status was performed on 5 studies22,26,29,30,34 that had sufficient data with a pooled value of 0.827 (95% CI , 0.760–0.894; I2 = 88.55%). For 1p19q status, a meta-analysis was performed on 4 studies22,26,34,35 that had sufficient data with a pooled value of 0.872 (95% CI, 0.789–0.954; I2 = 86.19%).
Derived aims and key findings of studies comparing IDH-mut and IDH wild-type LGG
Derived aims and key findings of studies examining 1p19q status of IDH-mut LGG
IDH status forest plot of included studies with an AUC.
1p19q status forest plot of included studies with an AUC.
The QUADAS-2 score showed low bias and high applicability (see Online Fig 2 for individual studies). The radiomic-specific RQS average score was low, with a mean of 10 (range, 2–14). On average, the RQS was 29% (range, 6%–39%) of the highest possible score. There were no studies that reported on cost-effectiveness, imaging used on phantom models, a prospectively validated radiomic signature in an appropriate clinical trial or performed clinical utility statistics (beyond just discussion of uses).21 Further details are provided in Online Table 3.
DISCUSSION
The systemic literature review found that the highest classifier for IDH status was conventional radiomics with CNN deep learning–derived features, which achieved an AUC = 0.95 (sensitivity of 94.4%, specificity of 86.7%).27 For classification of 1p19q status, conventional texture-based radiomics was optimal, with an AUC = 0.96 (sensitivity of 90%, specificity of 89%).34
Segmentation had manual components in both studies28,32 and was generally performed by trained personnel and approved by neuroradiologists or neurosurgeons. Manual segmentation is time-consuming, resource-intensive and introduces interobserver variability. Automation of segmentation is being actively progressed by the Brain Tumour Segmentation Challenge, and ongoing improvements have the potential to address the limitations of manual segmentation and thus improve the accuracy and efficiency of radiomic methods.36⇓-38 For the whole tumor, the 2018 winning team achieved a Sørensen–Dice coefficient of 0.88, in which a value of 1 represents perfect consistency between manual (ground truth) and automated segmentation.39
For IDH status, the literature indicates that a standard sequence image acquisition, use of texture-based features (most common being gray-level co-occurrence matrix,23-25,27-29,34,40 followed by the gray-level run-length matrix25,27⇓-30,34,40) with deep learning–derived features, and an SVM machine learning model may result in an optimal radiomic pipeline. One study classified solely using texture-based radiomic features and achieved an AUC = 0.79.34 Integration of deep learning with radiomic features did not increase the AUC in 1 study22 but produced the highest AUC = 0.95 in another study.27 Features derived from qualitative visual inspection (Visually Accessible Rembrandt Images; VASARI) did not increase the AUC compared with just radiomic features.34 Four studies examined multiparametric imaging.23,25,29,30 The entropy (randomness of voxel intensities) feature derived from ADC images was significantly different between IDH-mut and IDH wild-type LGG,23 suggesting that heterogeneity of ADC values may be helpful in predicting IDH status. Nevertheless, while integration of diffusion/perfusion imaging showed improved classification in 3 studies,25,29,30 ultimately it was not superior to using standard sequences with a different radiogenomic pipeline.28
For 1p19q status, the literature indicates that standard image sequences, use of texture-based features (the most common being grey-level run length matrix26,28,34,35 followed by gray-level co-occurrence matrix26,28,34), and a linear SVM machine learning model may result in an optimal radiomic pipeline. The highest AUC = 0.96 was achieved solely using texture-based radiomic features.34 Clinical and imaging-feature (such as age, sex, and the presence of bleeding or enhancement) integration did not improve the classification performance,38 nor did solely examining visually-created features.34 Deep learning feature integration with radiomic features increased classification performance; however, solely examining deep features was superior.22 The best-performing ML model classifier was achieved by a linear SVM.28
For studies included in the meta-analysis, there was high heterogeneity, given the variation in the unique elements of each radiomic pipeline. Heterogeneity is inevitable with any meta-analysis; however, acceptable levels may be a Higgins I2 of 0%–40%.41 The meta-analysis found 88.55% and 86.19% heterogeneity for IDH and 1p19q status, respectively. Although the QUADAS-2 showed low bias and high applicability, the radiomic-specific RQS assessment showed an overall inadequate clinical applicability of studies, identifying issues, including a lack of cost-effectiveness analysis, clinical utility statistics, or prospective validation. This is consistent with other neuro-oncologic radiomic studies in the literature.21 The RQS has some limitations, however. For example, greater emphasis is placed on the image-acquisition parameters12 than on the image-normalization process (making the voxel, section thickness, and matrix size similar among MR imaging scans), despite the latter being important for optimal translation into multi-institutional contexts. Of note, a perceived advantage of the AI algorithms is greater objectivity and thus a more consistent diagnosis, but this has yet to be convincingly proved in the literature.42
Classification of LGG for IDH status followed by further classification of 1p19q status (when IDH-mut) will have multiplicative effects. There was a sensitivity of 94.4% and specificity of 86.7%27 for IDH status, with a sensitivity of 90% and specificity of 89%34 for 1p19q. Thus, by using multiplication, we can find the maximum literature prediction of 1p19q status in an IDH-mut LGG to have a sensitivity of 85.0% (94.4% × 90%) and a specificity of 77.2% (86.7% × 89%). The conventional radiogenomic pipelines assume that the features assessed are independent, though they are not. For example, to take an example from the visual-feature literature, ill-defined tumor margins have been correlated with IDH wild-type LGG,43 but if the tumor is IDH-mut, it is more likely 1p19q-codeletion.10 There is also uncertainty regarding the interaction between radiomic and conventional visual MR imaging features. For example, if the T2-FLAIR mismatch sign is present, the literature would suggest that this can predict an IDH-mut 1p19q-intact glioma with greater confidence than radiomics.8,44 Yet, when these conventional features with the greatest predictive value are absent, one could expect that radiomics would predict the genotype better than other conventional MR imaging features. Thus, optimal classification may be achieved using a combination of conventional and radiomic features.
Acceptance of AI into clinical practice remains an issue. Much of the literature on integration is opinion-based,45⇓-48 and research related to understanding challenges is in its early stages.49⇓-51 Acceptance by patients also remains an issue; a recent study by Palmisciano et al52 found that only 66.3% of patients found it acceptable for AI to be used during imaging interpration.53 Issues raised by patients include distrust, lack of knowledge, a lack of personal interaction, questions about the efficacy of the AI algorithm, and the importance of being properly informed of its uses.54 Similar relevant issues were identified by a computer science literature review55 on human-AI interaction, such as task allocation, lack of knowledge and/or trust, incorrect use due to confusion, and integration issues due to a potentially radically different work practice.
Future directions for integration into the clinical sphere may come in the form of examining the nonmedical sphere,55 given successful implementation in other fields such as failure detection in truck engines and welding robots.56 One specific issue is that some AI programs used were developed in-house and may not be readily available to other institutions; important next steps include comparisons between programs and subsequent validation on larger external cohorts. There is also a lack of clinical trials assessing the integration of radiomic analysis into clinical practice,44 which was confirmed on our RQS assessment. Guidelines have recently been developed to address these issues, which may provide a framework for integration. For example, Microsoft has recently released a set of 18 general principles for integration into systems, such as explaining to the user (clinician) what the AI algorithm can do, how well it can be done and making it clear how it is performed.57 A thinking paradigm that may solve this is treating radiomic analysis as a new intervention or drug and applying ideas from existing protocols such as Phase I–IV clinical trials.58 The Food and Drug Administration has also recently released guidelines for AI integration into health care systems.59 Given that radiomic analysis is rapidly progressing and combining AI with standard radiologist assessment may show superior outcomes, there needs to be greater effort to translate findings into an interpretable format for clinical radiology.
CONCLUSIONS
The greatest classifier of IDH status in LGG was achieved with conventional radiomics in combination with convolutional neural network–derived features, providing a sensitivity of 94.4% and specificity of 86.7% (AUC = 0.95). Optimal classification of 1p19q status occurred using texture-based radiomics, with a sensitivity of 90% and a specificity of 89% (AUC = 0.96). The literature is limited by the use of manual segmentation, suboptimal study design, and the lack of translational work to integrate radiogenomic analysis into clinical practice.
Footnotes
Disclosures: Arian Lasocki—RELATED: Grant: Peter MacCallum Cancer Foundation, Comments: Arian Lasocki was supported by a Peter MacCallum Cancer Foundation Discovery Partner Fellowship, providing clinical backfill to allow dedicated research time.* *Money paid to the institution.
Indicates open access to non-subscribers at www.ajnr.org
References
- Received May 2, 2020.
- Accepted after revision August 17, 2020.
- © 2021 by American Journal of Neuroradiology