Abstract
BACKGROUND AND PURPOSE: Co-occurrence of local anisotropic gradient orientations (COLLAGE) is a recently developed radiomic (computer extracted) feature that captures entropy (measures the degree of disorder) in pixel-level edge directions and was previously shown to distinguish predominant cerebral radiation necrosis from recurrent tumor on gadolinium-contrast T1WI. In this work, we sought to investigate whether COLLAGE measurements from posttreatment gadolinium-contrast T1WI could distinguish varying extents of cerebral radiation necrosis and recurrent tumor classes in a lesion across primary and metastatic brain tumors.
MATERIALS AND METHODS: On a total of 75 gadolinium-contrast T1WI studies obtained from patients with primary and metastatic brain tumors and nasopharyngeal carcinoma, the extent of cerebral radiation necrosis and recurrent tumor in every brain lesion was histopathologically defined by an expert neuropathologist as the following: 1) “pure” cerebral radiation necrosis; 2) “mixed” pathology with coexistence of cerebral radiation necrosis and recurrent tumors; 3) “predominant” (>80%) cerebral radiation necrosis; 4) predominant (>80%) recurrent tumor; and 5) pure tumor. COLLAGE features were extracted from the expert-annotated ROIs on MR imaging. Statistical comparisons of COLLAGE measurements using first-order statistics were performed across pure, mixed, and predominant pathologies of cerebral radiation necrosis and recurrent tumor using the Wilcoxon rank sum test.
RESULTS: COLLAGE features exhibited decreased skewness for patients with pure (0.15 ± 0.12) and predominant cerebral radiation necrosis (0.25 ± 0.09) and were statistically significantly different (P < .05) from those in patients with predominant recurrent tumors, which had highly skewed (0.42 ± 0.21) COLLAGE values. COLLAGE values for the mixed pathology studies were found to lie between predominant cerebral radiation necrosis and recurrent tumor categories.
CONCLUSIONS: With additional independent multisite validation, COLLAGE measurements might enable noninvasive characterization of the degree of recurrent tumor or cerebral radiation necrosis in gadolinium-contrast T1WI of posttreatment lesions.
ABBREVIATIONS:
- COLLAGE
- co-occurrence of local anisotropic gradient orientations
- CRN
- cerebral radiation necrosis
- Gd-C
- gadolinium-contrast
- RT
- recurrent tumor
- TCIA
- The Cancer Imaging Archive
Currently >200,000 patients in the United States annually undergo chemoradiation as a standard-of-care treatment in primary and metastatic brain tumors.1 Following chemoradiation treatment, these patients typically undergo regular MR imaging (usually comprising T1WI, T2WI, FLAIR) for monitoring signs of tumor recurrence. A major clinical challenge in evaluating these posttreatment MR images is the differentiation of these lesions as recurrent tumor (RT) or cerebral radiation necrosis (CRN).2 CRN is an irreversible radiation-induced injury caused by aggressive radiation treatment and is challenging to diagnose on conventional MR imaging due to its close visual resemblance to tumor recurrence. The differentiation is further complicated by the simultaneous presence of varying proportions of CRN and recurrence/residual tumor confounding the diagnosis on imaging. Currently, the only definitive diagnosis of CRN rather than RT is via surgical intervention, followed by extensive histopathologic evaluation for establishing the extent of CRN or tumor recurrence in a lesion. On the basis of the extent of CRN intermixed with tumor, lesions can be characterized on histopathology as “pure” CRN (complete absence of tumor tissue), “predominant” CRN (>80% CRN), predominant RT (>80% tumor, <20% CRN), and “mixed” CRN (between 30% and 70% CRN). The current criterion standard diagnostic test for evaluating lesions posttreatment is surgical resection followed by extensive pathologic evaluation. Existing advanced noninvasive imaging protocols (ie, MR spectroscopy and PET) are known have interreader variability and have reported poor specificity in distinguishing RT from CRN.3,4
Recently, a few radiomics (computational feature-extraction approaches) studies in conjunction with routinely available MR imaging sequences have attempted to capture lesion heterogeneity for survival prediction and response assessment in brain tumors.5 Specifically, gray-level co-occurrence matrix-based features from active tumor regions were found to be predictive of brain tumor survival by Sottoriva et al.18 Gray-level co-occurrence matrix-based features have also been shown to be discriminative of phenotypes in glioblastoma.19 It has been shown by Rathore et al20 that peritumoral radiomic signatures could predict recurrence in glioblastoma and have further implications in personalized radiation therapy planning. While several of these recent studies have shown success in using radiomic analysis for survival prediction, only a few studies4,21 have explored distinguishing posttreatment changes (ie, CRN and pseudoprogression) from tumor recurrence using radiomic analysis.
In Prasanna et al,6 we presented a new radiomic feature, co-occurrence of local anisotropic gradient orientations (COLLAGE), that computes entropy (quantitative measurement that captures the degree of disorder) in voxelwise gradient orientations on routine gadolinium-contrast (Gd-C) T1WI. Specifically, we demonstrated that the COLLAGE entropy feature allowed differentiation between predominant CRN and predominant RT, with elevated expression of COLLAGE (reflective of high disorder in intensity gradients) being associated with tumor, and lower COLLAGE values, with RN.2 However, our study7 and other studies8,9 that have previously attempted to distinguish CRN from RT on imaging have been limited to investigating cases that were histologically identified as either predominant CRN or predominant tumor. This limitation is because posttreatment brain tumor lesions are rarely pure and largely exhibit a heterogeneous appearance owing to the prevalence of both CRN and tumor (referred to as mixed pathology).
The objective of this study was to reliably characterize different lesion pathologies of CRN and RT on routinely acquired posttreatment Gd-C T1WI using radiomics. On the basis of our previous observations using COLLAGE in predominant CRN/RT cases,6 in this feasibility study, we sought to investigate whether COLLAGE measurements are capable of distinguishing extreme (pure) from mixed pathologies for CRN and RT.
Specifically, in this study, we explored the association of COLLAGE measurements on posttreatment Gd-C T1WI with the extent of CRN and recurrent tumors across a cohort of 75 patients histologically confirmed and treated for nasopharyngeal carcinoma, primary, and metastatic brain tumors. Instances of pure CRN were obtained from patients with nasopharyngeal carcinoma10; CRN is an adverse effect of radiation in nasopharyngeal carcinoma because brain is a bystander during treatment. The manifestation of CRN in nasopharyngeal carcinoma, unlike in brain tumors, is unadulterated (pure) because there is no known malignant tumor presence in these brain lesions. Additionally, treatment-naïve brain tumor MR imaging from aggressive brain tumors (ie, grade IV glioblastoma) represents instances of pure tumor on imaging.
We investigated the following: 1) if and how first-order statistics (mean, median, skewness, and kurtosis) of COLLAGE measurements differ across different grades of pure, predominant, and mixed CRN and recurrent tumor in primary and metastatic tumors, and 2) whether these statistics provide improved discrimination across different pathologies of RT and CRN than using just MR imaging intensities alone.
Materials and Methods
Study Population
For this study, we accrued imaging scans of patients who had been diagnosed and treated for primary/metastatic brain tumors and nasopharyngeal carcinoma. The studies of patients with brain tumor were collected at the University Hospitals, Cleveland (site 1) and University of Texas Southwestern (site 2), while the nasopharyngeal carcinoma studies were obtained from the Tuen Mun Hospital, Hong Kong (site 3), with all cohorts accrued between 1990 and 2014.
Preoperative MRIs of subjects with glioblastoma used under the “pure tumor” category were made available for public download from The Cancer Imaging Archive (TCIA). TCIA is an open archive of cancer-specific medical images and associated clinical metadata established by the National Cancer Institute and collaborating institutions in the United States. A total of 10 MR imaging studies were randomly chosen from the TCIA cohort to be used as controls for pure tumor cases to maintain class balance across all categories.
A total of 75 studies were histologically confirmed with different degrees of CRN and tumor (categorized as pure, predominant, or mixed) in a lesion, details of which are provided in the Table. Inclusion criteria for studies across the 3 sites were the following: 1) the availability of 1.5 or 3T Gd-C T1WI, and 2) the pathology specimen obtained by resection or by at least 2 biopsies via stereotactic guidance for disease confirmation.
Patient cohorts with corresponding diagnosis as identified by an expert neuropathologist on postsurgical specimens
For sites 1 and 2, following the standard dose of concomitant radiation and chemotherapy, the patients who presented with suspicious posttreatment lesion artifacts, indicative of CRN or RT, were identified. Forty-two cases were accrued from site 1, consisting of 22 primary tumors identified as 12 predominant tumors and 10 cases of predominant CRN, and 20 metastatic tumors identified as 12 predominant tumors and 8 cases of predominant CRN. Similarly, 10 studies were accrued from site 2, with 7 primary and 3 metastatic tumors. Of the 7 cases of primary tumors, 4 had predominant tumor, 1 case was predominant CRN, and 1 case each had mixed pathologies of 30% and 75% CRN, respectively. The 3 metastatic cases consisted of 1 case each of predominant tumor, predominant CRN, and mixed pathologies.
Site 3 consisted of patients with pure CRN (n = 13) originally diagnosed with nasopharyngeal carcinoma who were symptomatic of CRN in the temporal lobe. Patients were treated with a standard dose of radiation therapy, 66–70 Gy in 33–35 fractions for radical treatment of nasopharyngeal carcinoma. The medial temporal lobe received nearly 100% of the prescribed dose to the nasopharynx. MR images were obtained for patients who developed CNS symptoms or as a part of the regular work-up for suspected local recurrence of nasopharyngeal carcinoma.
Studies from site 1 have previously been used in the initial development of the COLLAGE descriptor.5 Similarly, studies from site 2 have been used as an independent validation set by Prasanna et al.6
All MR images were acquired in axial sections with a 1.5T or 3T scanner. We acquired T1-weighted postcontrast images with the following parameters: For sites 1 and 2, the mean TR and TE were 250 and 2.48 ms, respectively. For site 3, the corresponding values were 620 and 20 ms, respectively.
Confirmation of Disease Presence
For sites 1 and 2, the patient cohort was selected by performing a retrospective review of neuropathology in all patients with brain tumors who underwent a surgical intervention for a recurrent or progressive Gd-C T1WI–enhancing lesion identified during follow-up at 9 months (or later) after the initial radiation therapy. Follow-up MR images within 0–21 days before the second resection or multiple biopsies (for disease confirmation) were used for analysis. Histology was rereviewed by a neuropathologist (M.C. at site 1 and K.H. at site 2) blinded to the original diagnosis and type of RT, to quantify the percentage of CRN and RT. Histopathologic diagnosis was based on World Health Organization criteria, which included factors like the degree of pleomorphism, mitoses, and vascular proliferation among others.11,12
For site 3, disease confirmation was obtained by histologic diagnosis by an expert who analyzed the presence of pathologic features of CRN such as fibrinoid and coagulative necrosis. For limited cases in which confirmation was not feasible using histologic analysis, radiographic monitoring on follow-up MR images was used for disease confirmation.
Preprocessing
To account for variability in pixel resolutions across the studies, we resampled every MR imaging slice to a uniform pixel spacing of 1 × 1 mm2. Similarly, every MR imaging volume was interpolated to a 3-mm slice thickness. We then corrected every study for intensity nonstandardness, which refers to the inherent drift in MR imaging signal intensities during acquisition, an artifact causing image-intensity values within tissue-specific compartments to vary across sites, scanners, and even within multiple repeat scans of a single patient. Correction for intensity nonstandardness was implemented using the approach as described in Madabhushi and Udupa13 and implemented in Matlab R2016a (MathWorks, Natick, Massachusetts). Specifically, we used a piecewise continuous histogram-matching method (bins = 255, points = 64) for normalizing the multi-institutional MR imaging studies. Additional preprocessing involving skull stripping was performed14 via the skull-stripping module in 3D Slicer (http://www.slicer.org).15 Then, lesions were annotated on Gd-C T1WI by 2 radiology experts (L.W., A.S.) on the slice that presented the maximum lesion conspicuity. The 2 radiologists (L.W., A.S.) worked in conjunction to establish a consensus to define an ROI that was used for further analysis.
Feature Extraction and Statistical Analysis
Within the ROI identified by consensus across the expert radiology readers on Gd-C T1WI, COLLAGE measurements were extracted on a per-pixel basis for all the pixels within the ROI. Briefly, COLLAGE involves extracting the dominant gradient orientation along the X and Y directions for every pixel via principal component analysis.16 A co-occurrence matrix is then computed for every pixel within the neighborhood to capture co-occurring arrangements of the dominant gradient orientations. Thirteen different second-order measurements (such as energy, entropy, and variance) are then computed from this co-occurrence matrix of the dominant gradient orientations. We chose to focus on entropy because it captures the disorder of pixel gradient orientations on a per-pixel basis. In Prasanna et al,6 we showed that the entropy values for the localized orientations were high for tumor, while the values were low for benign pathologies (ie, CRN). A detailed description of the algorithm and methodology for computing 3D COLLAGE can be found in Prasanna et al.6 For our analysis, every image voxel within the enhancing lesion was assigned a COLLAGE entropy value. We subsequently extracted 4 different first-order statistics of the features (mean, SD, skewness, and kurtosis) across all the voxels within the most conspicuous lesion per study. For each of the 4 statistics, the range of values was rescaled between 0 and 1. Each study was hence represented by a 4 × 1 feature vector, comprising the 4 statistics of COLLAGE entropy within that ROI.
Comparison of COLLAGE Radiomic Features across Different Grades of Radiation Necrosis and Recurrent Tumor in Primary and Metastatic Tumors
Statistics of COLLAGE values (mean, SD, skewness, and kurtosis) were compared for the pure CRN pathology against mixed and predominant CRN/recurrent tumor pathologies, using the Wilcoxon rank sum test, independently across primary and metastatic tumors. We included COLLAGE statistics from the TCIA glioblastoma studies to represent the pure tumor class, while comparing pathologies within the primary tumor cohort. The statistics obtained from COLLAGE were also compared with those obtained from average Gd-C T1WI intensity values across different CRN and recurrent tumor pathologies.
Classification Analysis to Distinguish CRN from RT Using Pure and Predominant CRN and Tumor COLLAGE Features
A random forest classifier was trained separately on primary and metastatic cases, in a leave-one-out cross-validation setting. First, COLLAGE features from only predominant CRN/tumor (from site 1) were used within the training cohort. Next, we incorporated the pure CRN signatures (from site 3) into the training set and repeated the classifier analysis separately for primary and metastatic tumors. It was ensured that all the folds within leave-one-out cross-validation contained pure CRN studies within the training set. Receiver operating characteristic curves were obtained for the training set analysis along with the corresponding areas under the curve.
We used the classifier trained with both pure and predominant COLLAGE features and evaluated its performance on the predominant cases on the held-out cases from site 2 (n = 10). Three studies from site 2 that had mixed degrees of CRN and tumor were excluded from the test set because the samples in the training cohort were not exposed to mixed degrees of CRN. However, the statistics obtained from the COLLAGE values for these studies were compared with those obtained from the training cohort and were reported.
Results
Comparison of COLLAGE Radiomic Features across Different Grades of Radiation Necrosis and Recurrent Tumor in Primary and Metastatic Tumors
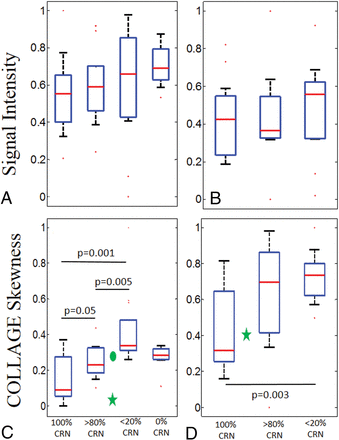
The original Gd-C T1WI intensity values were not found to be statistically different across predominant CRN, predominant tumor, pure CRN, and pure tumor groups for the primary cohort (Fig 1A). While mean, SD, and kurtosis statistics of COLLAGE did not demonstrate significant differences, the corresponding COLLAGE skewness values were found to be statistically significantly different (P < .05) across the 3 categories (excluding pure tumor in which the results were not statistically significantly different from predominant CRN) as shown in Fig 1C, with higher skewness values suggesting a skewed distribution toward elevated COLLAGE entropy. Most interesting, for primary tumors, COLLAGE skewness values were lowest for pure CRN with a mean of 0.15 ± 0.12 and 0.25 ± 0.09 for predominant CRN, 0.42 ± 0.21 for predominant recurrence, and 0.27 ± 0.07 for pure tumor, respectively. The values were statistically significantly different between predominant CRN and predominant tumor recurrence with P = .005, between pure CRN and predominant tumor recurrence with P = .001, and between pure CRN and predominant CRN with P = .05.
Box-and-whisker plots showing the distributions of T1WI signal intensities and skewness of COLLAGE entropy values for pure and different grades of CRN for primary and metastatic cases, respectively. A and B, The signal intensities in primary and metastatic cases, respectively. C and D, The COLLAGE skewness values for the same cases. C, The green star and circle represent the cases from site 2 with 75% CRN and 30% CRN, respectively. D, The green star represents the case from site 2 with 50% CRN. P values for the pairs of groups exhibiting statistically significant difference are also shown.
Qualitative results of COLLAGE feature maps of pure CRN, predominant CRN, and predominant tumor reflecting corresponding changes in radiomic values are shown in Fig 2. COLLAGE values for both mixed pathology studies were found to lie between predominant CRN and tumor categories. The study with 75% CRN (the green star in Fig 1C) had a low skewness value of 0.013 compared with the one with 30% CRN (the green circle in Fig 1C) with a skewness value of 0.285.
Gd-C T1WI for CRN from nasopharyngeal carcinoma (A), predominant (>80%) CRN (B), predominant recurrent tumor (<20% CRN) (C), and glioblastoma (0% CRN) (D), and the corresponding COLLAGE entropy maps of the ROIs (E–H).
Similarly, for the metastatic cases, the intensity values (Fig 1B) and mean, SD, and kurtosis of COLLAGE features were found not to be statistically significantly different across the 3 groups. However, COLLAGE skewness values (ranging between 0 and 1) were significantly different, with the lowest values for pure CRN with a mean of −0.41 ± 0.22 and 0.61 ± 0.33 for predominant CRN, and 0.72 ± 0.14 for predominant recurrence, respectively. COLLAGE values were statistically significantly different only between pure CRN and predominant recurrence with P = .003, and not across predominant and pure CRN. The study in site 3 with 50% CRN (green star in Fig 1D) had a skewness value of 0.4.
Classification Analysis to Distinguish CRN from RT Using Pure and Predominant CRN and Tumor COLLAGE Features
For primary brain tumor cases, the area under the curve for the classifier, using COLLAGE features from both pure CRN and predominant studies in the training cohort, was found to be 0.67, while for metastatic tumors, the area under the curve was 0.66. The corresponding accuracies at the optimal operating point were found to be 68.2% and 65%, respectively. However, the area under the curve using only the COLLAGE features from predominant studies was found to be 0.64 for primary and 0.56 for metastatic tumors. The corresponding accuracies at the optimal operating point were found to be 59.1% and 65%, respectively. On the independent test set at site 2, a total of 4 of 5 primary tumor cases with predominant CRN/tumor were correctly classified. For the metastatic tumor cases, 1 of the 2 cases was correctly classified.
Discussion
COLLAGE, a recently developed radiomic feature, has previously been shown to be effective in distinguishing predominant recurrent brain tumors from cerebral radiation necrosis on MR imaging6 and also in predicting a pathologic complete response to neoadjuvant chemotherapy in breast cancer.17 In this work, we attempted to interrogate differences in COLLAGE values on posttreatment Gd-C T1WI across different posttreatment pathologies in brain tumors by associating the extent of CRN and tumor with the corresponding expression levels of COLLAGE. We present our findings of associations of COLLAGE features across pathologically proved pure, predominant, and mixed categories of CRN and tumor.
While in the metastatic cohort, statistically significant differences were observed only between pure CRN and predominant recurrence, COLLAGE values in the primary cohort were statistically significantly different between predominant CRN and predominant recurrence, pure CRN and predominant recurrence, and pure CRN and predominant CRN. Over and above the results reported in Chao et al,2 the findings in this work suggest that COLLAGE may potentially characterize the spectrum of pathologies of CRN and RT on posttreatment Gd-C T1WI, as confirmed on the corresponding histopathology.
Furthermore, our results show that COLLAGE entropy values were skewed toward higher values for the predominant tumor cases compared with the pure CRN or predominant CRN for both primary and metastatic tumors (Fig 1). The difference in skewness may be attributed to the low COLLAGE values in lesions having lower concentrations of CRN, likely characterizing relatively inactive necrotic tissue. Given that tumors have increased heterogeneity,18 it is intuitive that COLLAGE features tend to be overexpressed, resulting in higher skewness, while radiation necrosis, which tends to have a more coherent microarchitecture, results in a muted COLLAGE response and consequently lower skewness values.
The reason for lower COLLAGE skewness in predominant RT than in pure RT is likely the presence of more heterogeneous tissue pathologies in the predominant RT group. Predominant RT often exhibits tissue heterogeneity owing to the varying presence of radiation-induced vascular hyalinization, telangiectasia, and zonal necrosis. This tissue heterogeneity is possibly manifested on the imaging scale and may therefore be captured by the skewness of COLLAGE entropy. A similar trend was observed in the variance of the skewness values in the boxplots, with low variance in COLLAGE observed in pure tumor compared with predominant RT.
Our results further demonstrate that incorporating COLLAGE features from pure CRN in the training set resulted in improved classification performance of the predominant CRN/RT, compared with using COLLAGE features from the predominant CRN/RT alone. These findings are consistent from a machine learning perspective, wherein an ideal classifier is expected to be exposed to the entire spectrum of cases—that is, a learning set comprising features corresponding to “pure CRN and no cancer” and mixed classes comprising co-existing CRN and cancer.
There were a few limitations of this feasibility study. Our cohort was retrospectively acquired as a part of a study that was concluded in 2016. Because we limited the analysis to pathologically proved grades of CRN/RT, the sample size was relatively small. While our retrospective cohort contained pathologically proved CRN and RT cases, it lacked cases that were histologically confirmed to be pure metastatic brain tumor. Hence, we could not perform a similar analysis on the metastatic cases, as was performed on the cases with primary brain tumors. Furthermore, differences in radiation therapy protocols in CNS tumors (both primary and metastatic) and nasopharyngeal carcinoma were not considered in the analysis. The analysis was limited to only Gd-C T1WI in this study and did not consider other routine or advanced imaging protocols. No independent large-scale validation of the findings was performed as a part of this study.
Conclusions
In this feasibility analysis, we presented the initial results of using a new radiomic feature, COLLAGE, to capture the extent of cerebral radiation necrosis and recurrent tumor on posttreatment Gd-C T1WI. We identified associations of COLLAGE features with the extent of CRN and RT on Gd-C T1WI on a unique cohort of patients histologically confirmed and treated for nasopharyngeal carcinoma, and primary and metastatic brain tumors. These preliminary findings suggest that COLLAGE may be potentially capturing subtle differences across different pathologic categories of RT and CRN (pure, predominant, and, to some extent, mixed) on posttreatment Gd-C T1WI. Learning such signatures, following extensive multisite validation, may, in the future, help in improved discrimination of CRN and RT, which still remains an extremely challenging clinical problem in neuro-oncology. In future work, we intend to prospectively validate our preliminary findings on a larger, multi-institutional cohort.
Footnotes
Disclosures: Leo Wolansky—UNRELATED: Other: Guerbet grant to the University of Connecticut.* Anant Madabhushi—UNRELATED: Board Membership: Inspirata; Consultancy: Inspirata; Grants/Grants Pending: Inspirata, PathCore, Comments: involved in 3 National Cancer Institute grants with Inspirata and 1 U24 grant (National Cancer Institute) with PathCore*; Patents (Planned, Pending, or Issued): Patents are licensed to Inspirata through Rutgers and Case Western*; Royalties: Royalties are paid out to Case Western and Rutgers by Inspirata and Elucid Bioimaging*; Stock/Stock Options: Elucid Bioimaging, Inspirata. Pallavi Tiwari—RELATED: Grant: The Dana Foundation, Ohio Third Frontier, I Corps.* *Money paid to the institution.
This work was supported by the National Cancer Institute of the National Institutes of Health under award Nos. 1U24CA199374–01, R21CA179327–01, R21CA195152–01; the National Institute of Diabetes and Digestive and Kidney Diseases under award No. R01DK098503–02; the Department of Defense Prostate Cancer Synergistic Idea Development Award (PC120857); the Department of Defense Lung Cancer Idea Development New Investigator Award (LC130463); the DOD Prostate Cancer Idea Development Award (W81XWH-15-1-0558); the Case Comprehensive Cancer Center Pilot Grant, VelaSano Grant, from the Cleveland Clinic; and the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Indicates open access to non-subscribers at www.ajnr.org
References
- Received June 26, 2018.
- Accepted after revision December 13, 2018.
- © 2019 by American Journal of Neuroradiology