Abstract
BACKGROUND AND PURPOSE: Conventional MR imaging explains only a fraction of the clinical outcome variance in multiple sclerosis. We aimed to evaluate machine learning models for disability prediction on the basis of radiomic, volumetric, and connectivity features derived from routine brain MR images.
MATERIALS AND METHODS: In this retrospective cross-sectional study, 3T brain MR imaging studies of patients with multiple sclerosis, including 3D T1-weighted and T2-weighted FLAIR sequences, were selected from 2 institutions. T1-weighted images were processed to obtain volume, connectivity score (inferred from the T2 lesion location), and texture features for an atlas-based set of GM regions. The site 1 cohort was randomly split into training (n = 400) and test (n = 100) sets, while the site 2 cohort (n = 104) constituted the external test set. After feature selection of clinicodemographic and MR imaging–derived variables, different machine learning algorithms predicting disability as measured with the Expanded Disability Status Scale were trained and cross-validated on the training cohort and evaluated on the test sets. The effect of different algorithms on model performance was tested using the 1-way repeated-measures ANOVA.
RESULTS: The selection procedure identified the 9 most informative variables, including age and secondary-progressive course and a subset of radiomic features extracted from the prefrontal cortex, subcortical GM, and cerebellum. The machine learning models predicted disability with high accuracy (r approaching 0.80) and excellent intra- and intersite generalizability (r ≥ 0.73). The machine learning algorithm had no relevant effect on the performance.
CONCLUSIONS: The multidimensional analysis of brain MR images, including radiomic features and clinicodemographic data, is highly informative of the clinical status of patients with multiple sclerosis, representing a promising approach to bridge the gap between conventional imaging and disability.
ABBREVIATIONS:
- DD
- disease duration
- EDSS
- Expanded Disability Status Scale
- IQR
- interquartile range
- MAE
- mean absolute error
- ML
- machine learning
- MS
- multiple sclerosis
- WBV
- whole-brain volume
MR imaging is firmly established as a fundamental tool for the diagnosis1 and monitoring2 of multiple sclerosis (MS), with MR imaging features commonly used as surrogate markers of disease activity in both clinical trials3 and routine clinical practice.4 However, conventional MR imaging measures (ie, the number, volume, and gadolinium enhancement of WM lesions) explain only a small fraction of the diversity of clinical outcomes in MS,5 with this mismatch traditionally referred to as the “clinicoradiologic paradox.”6 The reasons for this apparent dissociation are manifold, embracing the difficulty to both define and measure clinical disability and the inability of conventional MR imaging to exhaustively characterize CNS structural and functional modifications in MS.6 From a clinical standpoint, the Expanded Disability Status Scale (EDSS) score remains the most widely used outcome measure to assess MS-related disability in clinical trials.7 From a neuroimaging perspective, however, many research studies have attempted to address these blind spots, leveraging advanced MR imaging techniques to identify clinically relevant disease biomarkers (eg, brain global and regional atrophy,8 spinal atrophy,9 cortical lesions,10 microstructural damage of normal-appearing white matter, normal-appearing white matter,11 and GM12 changes of structural and functional brain networks13) that have the potential to bridge the gap between MR imaging and disability in MS and are progressively being integrated into clinical scenarios.
Of note, technical advances and the implementation of imaging guidelines2,14 have led to the widespread availability of good-quality clinical scans, including isotropic sequences suitable for volumetric quantifications.8 Furthermore, new promising connectomic approaches have shifted the emphasis from the sole quantification of total lesion burden to the functional consequences of WM damage in terms of brain network economy,13 which can also be coarsely inferred from macroscopic T2 lesions.15 Finally, the diffusion of radiomics has considerably augmented the amount of potentially meaningful information extractable from clinical images,16 with machine learning (ML) methods providing the means for more flexible modeling of high-dimensional neuroimaging data sets compared with traditional statistical approaches.17
Given this background, we aimed to conceptually address the “clinicoradiologic paradox” in MS by evaluating machine learning models for EDSS score prediction on the basis of a systematic mapping of textural, volumetric, and macrostructural disconnection features derived from routine brain MR images. The results were validated by external testing on a separate data set obtained from a second institution.
MATERIALS AND METHODS
Subjects
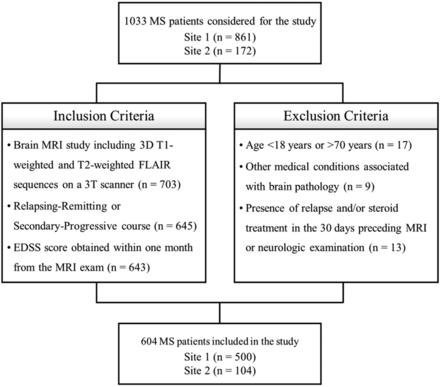
In this retrospective cross-sectional study, brain MR imaging studies of consecutive patients with an MS diagnosis revised according to the 2010 McDonald criteria18 and a relapsing-remitting or secondary-progressive19 course including 3D T1-weighted and T2-weighted FLAIR sequences were selected from the radiologic databases of 2 institutions: the MS Center of the University of Naples “Federico II” (site 1) and the Human Neuroscience Department of the University of Rome “Sapienza” (site 2). All studies were performed between October 2006 and January 2020. Clinical disability was estimated using EDSS scores obtained within 1 month of the MR imaging. Exclusion criteria were as follows: younger than 18 years of age or older than 70 years of age; other pre-existing major systemic, psychiatric, or neurologic disorders; and the presence of a relapse and/or steroid treatment in the 30 days preceding the MR imaging (Fig 1).
Flow diagram showing inclusion and exclusion criteria.
The study was conducted in compliance with the ethical standards and approved by the local Ethics Committees, and written informed consent was obtained from all subjects according to the Declaration of Helsinki.
MR Imaging Data Acquisition
All MR images were acquired with a 3T scanner and included a 3D T1-weighted sequence (≤1-mm isotropic resolution) for volumetric analyses and a T2-weighted FLAIR sequence for quantifying total demyelinating lesion volume. Details about sequence parameters are provided in the Online Supplemental Data.
MR Imaging Data Processing
A flow chart summarizing the data-processing pipeline is available in Fig 2, while a complete description of all processing steps is provided in the Online Supplemental Data.
Workflow summarizing the main MR imaging data-processing and data-mining steps. Image illustrates the data set composition as well as the major steps performed for feature extraction, feature selection, and regression modeling. LL indicates lesion load; NAWM, normal-appearing white matter; ChaCo, change in connectivity; T1w, T1-weighted imaging.
Volumetric Analysis.
Demyelinating lesions were automatically segmented on FLAIR images using the Lesion Segmentation Tool (LST), Version 3.0.0 (www.statistical-modelling.de/lst.html) for SPM (http://www.fil.ion.ucl.ac.uk/spm/software/spm12). Lesion probability maps were used to fill lesions in T1-weighted images for subsequent processing steps and binarized to compute total lesion volume.
Filled T1-weighted volumes underwent the segmentation pipeline implemented in the Computational Anatomy Toolbox (CAT12.6; http://www.neuro.uni-jena.de/cat) in SPM to obtain an atlas-based parcellation into 114 brain regions defined according to the CAT12-adapted version of the Automated Anatomical Labeling atlas (https://github.com/muschellij2/aal).20 Furthermore, whole-brain volume (WBV), GM subregion ROIs (and corresponding volumes), and normal-appearing white matter masks were also derived from the segmentation procedure.
Finally, total intracranial volume was estimated using CAT12, and brain volumes (both WBV and GM regions) were transformed into z scores while adjusting for age, sex, and estimated total intracranial volume.
Connectivity Analysis.
Subject-wise, for each of the 116 GM cortical/subcortical regions defined in the Automated Anatomical Labeling atlas,20 a change in the connectivity score was computed using the Network Modification (NeMo) tool,15 representing an estimate of local structural disconnection caused by WM tract disruption, as inferred from the location and load of WM lesions.
Radiomics Analysis.
First-order and texture features were extracted from each segmentation-derived ROI (normal-appearing white matter and 114 GM regions) from the unfilled T1-weighted volumes using PyRadiomics, Version 3.0.21 Before the extraction, the images underwent standard preprocessing steps. An exhaustive description of the features obtainable by PyRadiomics is available in the official documentation (https://pyradiomics.readthedocs.io/en/latest/features.html).
Radiomics feature stability with respect to the MR imaging processing pipeline was tested on a subset of 30 randomly selected subjects, and only features with excellent stability (intraclass correlation coefficient ≥ 0.90) were retained for subsequent analyses.
Machine Learning
ML analyses were performed using the Weka data mining platform (Version 3.8.3; http://old-www.cms.waikato.ac.nz/∼ml/weka/)22 and scikit-learn Python package (https://scikit-learn.org/stable/index.html).23 Given the nature of the EDSS score, regression algorithms were used to develop predictive models, with several algorithms (ie, ridge regression, support-vector machine, random forest, and Gaussian process) investigated to assess differences in performance due to model architecture. A description of the ML algorithms is provided in the Online Supplemental Data.
The site 1 cohort was randomly split into training (80% of subjects) and test (20% of subjects) sets for model tuning and testing, respectively, while the site 2 cohort was exclusively used as an external test set. After data-preprocessing (details in the Online Supplemental Data), clinicodemographic (age, sex, disease duration [DD], disease course), textural, and other MR imaging–derived (T2 lesion volume, WBV, and change in connectivity scores for each GM region) variables underwent multiple feature-selection steps on the training set. First, low variance (0.01 threshold) and highly colinear (>0.8) features were removed. Then, LASSO regression (https://www.lasso.io/), using the EDSS score as the dependent variable, was used to remove features whose coefficients shrank to zero. Finally, the Weka correlation–based subset evaluator was used to identify the best feature subset for EDSS prediction.
The resulting data set was used to train the 4 ML regression algorithms, whose tuning and initial performance evaluation were performed using 10-fold cross-validation in the training cohort (80% of site 1 data). Each final model was then assessed on the previously unseen cases of both the internal (remaining 20% of site 1 data) and external (site 2 data) test sets.
As an ancillary analysis, the models were retrained using clinicodemographic features exclusively, to indirectly assess the incremental benefit provided by imaging-derived measures.
Statistical Analysis
Statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS, Version 25.0; IBM) with a significance level α = .05. Between-site differences in terms of clinicodemographic variables were tested using the Student t test (age and disease duration), Fisher exact test (sex and disease course), and median test (EDSS), respectively. The effect of different ML algorithms on model performance was tested using 1-way repeated measures ANOVA with absolute errors as the dependent variable, including post hoc tests to compare each pair of predictive models, Bonferroni-corrected for controlling the family-wise error rate.
RESULTS
Subjects
A total of 500 patients with MS were selected from site 1 (428 relapsing-remitting, 72 secondary-progressive; mean age, 37.5 [SD, 10.9] years; male/female ratio = 151:349; mean disease duration = 9.3 [SD, 8.1] years). After the data set split, 400 subjects from site 1 constituted the training set, and 100 subjects, the internal test set. From site 2, one hundred four demographically and clinically comparable patients (84 relapsing-remitting, 20 secondary-progressive; mean age, 38.3 [SD, 9.8] years; male/female ratio = 24:80; mean disease duration = 9.2 [SD, 8.5] years) were included in the external test set.
The median EDSS score was 2.5 (interquartile range [IQR] = 2.0–4.0) and 2.0 (IQR = 1.5–4.0) for patients from sites 1 and 2, respectively (P = .03). In the overall study population, patients with secondary-progressive MS showed a higher EDSS score (median, 5.5; IQR = 4.5–6.0) than those with relapsing-remitting MS (median, 2.5; IQR = 2.0–3.5) (P < .001, accounting for age, sex, and disease duration). Demographic and clinical variables of all subjects included in the study are reported in Table 1.
Clinicodemographic characteristics of the studied population, along with MR imaging–derived global brain volumesa
MR Imaging Data Analyses and ML Predictive Models
For each participant, MR imaging–derived global (T2 lesion volume and WBV, also reported in Table 1) and regional (114 GM regions) brain volumes were computed, along with the change in connectivity scores corresponding to the 116 GM parcels of the Automated Anatomical Labeling atlas.
Furthermore, a total of 125,580 radiomics features were extracted from the 115 segmentation-derived ROIs (normal-appearing white matter and 144 GM regions), of which 43 were excluded as having nonexcellent reproducibility.
The feature-selection procedure, performed on the training cohort, identified 4907 low-variance and 99,312 highly colinear features. At LASSO regression, 21 features were selected, further reduced to 9 by the subset evaluator. These consisted of age and secondary-progressive course in addition to a subset of radiomic features (details in Table 2), which were then used to train the ML algorithms for EDSS score prediction. The trained model hyperparameters and ridge regression feature weights are available in the Online Supplemental Data.
Selected radiomics featuresa
Correlation coefficients (r) of the final models predicting EDSS scores in the 10-fold cross-validation in the training cohort ranged from 0.79 (R2 = 0.62, mean absolute error [MAE] = 0.66) for the random forest model to 0.80 (R = 0.64, MAE = 0.65) for ridge regression. On the internal test set, performances ranged from r = 0.73 (R = 0.54, MAE = 0.87) for Gaussian process regression to r = 0.74 (R2 = 0.55, MAE = 0.72) for ridge regression, while in the external test set, they ranged from r = 0.755 (R = 0.570, MAE = 1.155) for ridge regression to r = 0.799 (R2 = 0.638, MAE = 1.247) for Gaussian process regression (Table 3).
Machine learning predictive modelsa
There was a significant effect of the ML algorithm on the model performance in both the internal [F(1.82, 180.39) = 7.94] (P = .001) and external [F(1.70, 175.52) = 5.25] (P = .009) test sets. In particular, on the internal test set, Gaussian process regression performed significantly worse than all other algorithms (Bonferroni-corrected P ≤ .01), while support-vector machine regression performed significantly better than Gaussian process regression on the external test set (Bonferroni-corrected P < .001). Details of the pair-wise comparisons between different model performances are reported in the Online Supplemental Data.
As for the ancillary analysis, while it was clear that clinical features substantially contribute to EDSS prediction, with ridge regression and support-vector machine regression yielding the best overall results on the external test set (r = 813, MAE = 1.005 and r = 0.814, MAE = 0.945, respectively), models using these alone were much less consistent across the 3 data sets, with performance varying greatly on the basis of algorithm architecture (Online Supplemental Data).
DISCUSSION
In this study, we proved that predictive models based on textural features extracted from routine brain MR images, along with basic clinicodemographic data, correlate with clinical disability in patients with MS with high accuracy and intra- and intersite generalizability.
Since its earliest days, MR imaging research in MS has had the objective of unraveling the relationship between neuroradiologic imaging and clinical status.24 Indeed, several studies investigated the association between conventional MR imaging markers of MS pathology and EDSS, reporting correlation coefficients ranging from 0.15 to 0.6024 and nurturing the concept of a “clinicoradiologic paradox.”6 Through the years, many of the confounders sustaining this apparent contradiction have been addressed, with emphasis on a more specific characterization of CNS structural and functional modifications through advanced MR imaging techniques and a finer assessment of MS-related disability, including the evaluation of cognitive performance.5
Most interesting, a more recent study has dealt with this classic issue using a multivariate statistical analysis of local intensity patterns on conventional MR images of a small homogeneous sample of patients with MS, leading to promising results.25 The potential of ML in the analysis of MR imaging data in MS is also highlighted by another recent study using models based on FLAIR images and demographic information for the prediction of 2-year clinical disability and achieving a mean squared error of 3 (corresponding to a mean EDSS score error of 1.7).26 Furthermore, studies with a large number of subjects demonstrated the clinical relevance of automatic volumetric quantifications, systematically mapping brain anatomy at both global and regional levels on clinical MR images of patients with MS.27,28
In our work, we revisited the conventional MR imaging/clinical disability dissociation problem in the light of recent developments in the fields of radiomics and ML modeling, exploring the informative value of volumetric, macrostructural disconnection and textural features derived from routine MR images of a large multisite cohort of patients with MS. We found that ML models based on radiomics features extracted from specific brain regions, along with basic clinicodemographic data, are highly predictive of the EDSS score (r approaching 0.80, about 64% of shared variance), demonstrating excellent intra- and intersite generalizability (r ≥ 0.73, about 53% of shared variance). Of note, the ML algorithm had little effect on the predictive performance, with similar prediction errors across models, indicating substantial model-independence of our findings. Also, as per our ancillary analysis, while clinicodemographic variables alone were highly informative of patients’ clinical status, the inclusion of radiomics features in the models substantially increased the generalizability and stability across different ML algorithms, supporting the additional value of a holistic approach including a variety of data types/sources.
Although a meaningful comparison of effect sizes among studies is hindered by the variability of study design, sample size, and statistical methods, our study seemingly provides a sensible improvement compared with earlier works,6 with sample width and external validation across different sites further strengthening our results.
Most interesting, our findings confirm that signal intensity patterns as assessed by the quantitative texture analysis of conventional brain MR images encode clinically relevant information,25 apparently outperforming measures like volume or macrostructural disconnection in terms of shared variance with clinical disability. Indeed, textural features may capture subtle modifications of brain tissue microstructure, which are known to correlate with clinical status in patients with MS.12 Furthermore, the systematic mapping of different brain regions through atlas-based automatic segmentation of T1-weighted volumes may enhance radiomics analysis by adding anatomic specificity, with most informative features in our models extracted from areas whose pathologic modifications are known to impact the clinicocognitive performance (ie, prefrontal cortex,29 deep gray matter,27 and cerebellum30). Conversely, a simpler shape feature like volume, as well as the coarse estimation of GM structural disconnection as inferred by T2 lesion location, may represent less pathologically specific disease markers, therefore providing a minor contribution to explaining MS-related disability.
To date, few studies have explored the potential of radiomics in MS, mainly focusing on the analysis of WM lesions for diagnostic classification purposes,31,32 with alterations of brain tissue microstructure mostly characterized through advanced MR imaging techniques,33,34 which provide more neurobiologically interpretable results but require dedicated acquisitions that are difficult to implement in large-scale population studies. Nevertheless, the systematic radiomics analysis of conventional brain MR images may hide a huge unused potential, promising to exploit the maximum clinically meaningful information contained in neuroradiologic images, taking full advantage of a massive amount of clinical MR imaging data collected through the years.
Some limitations of the current study should be acknowledged. First, using EDSS as a measure of clinical severity has several shortcomings, including incomplete coverage of CNS domains, nonlinearity, low sensitivity, and inter- and intraobserver variability.35 However, despite these limitations and the availability of alternative rating scales, the EDSS is still considered the reference method to assess MS-related disability in both clinical trials and routine, therefore being more scalable to real-world scenarios.35 Furthermore, it is known that radiomics features may have instability due to variations in scanner and image-acquisition parameters.36 Nevertheless, this issue may be mitigated by the combined evaluation of basic clinicodemographic variables and other MR imaging–derived metrics with proved robustness (eg, automatic volumetric quantifications8) as suggested by the excellent generalizability of our models across different sequences and scanners. Finally, our work paves the way for future studies exploiting the proposed methodologic framework to predict longitudinal clinical outcomes, possibly providing a tool for effective prognostic stratification of patients with MS in clinical practice.
CONCLUSIONS
We demonstrated that the multidimensional analysis of routine brain MR images, including the systematic investigation of textural features in conjunction with basic clinicodemographic data, is highly informative of the clinical status of patients with MS. In the era of big data, this approach may represent a way of filling the gap between conventional imaging and clinical disability in MS.
Footnotes
Disclosures: Maria Petracca—UNRELATED: Travel/Accommodations/Meeting Expenses Unrelated to Activities Listed: travel sponsorship from Novartis. Nikolaos Petsas—UNRELATED: Consultancy: Istituto di Ricovero e Cura a Carattere Scientifico Istituto Neurologico Mediterraneo, Pozzilli, Italy; Employment: Sapienza University of Rome, Italy; Grants/Grants Pending: Onlus Sant’Andrea, Rome, Italy.* Carlo Pozzilli—UNRELATED: Board Membership: Merck, Biogen, Novartis, Bristol Myers Squibb, Almirall, Sanofi, Roche; Consultancy: Novartis; Grants/Grants Pending: Merck, Roche, Biogen*; Payment for Lectures Including Service on Speakers Bureaus: Almirall, Bayer, Biogen, Genzyme, Merck Serono, Novartis, Roche and Teva; Payment for Manuscript Preparation: Merck, Biogen, Roche; Payment for Development of Educational Presentations: Multiple Sclerosis Paradigm. Vincenzo Brescia Morra—UNRELATED: Consultancy: Biogen, Teva Pharmaceutical Industries, Genzyme, Merck, Novartis and Almirall; Payment for Lectures Including Service on Speakers Bureaus: Biogen, Teva Pharmaceutical Industries, Genzyme, Merck, Novartis, and Almirall. Patrizia Pantano—UNRELATED: Grants/Grants Pending: Fondazione Italiana Sclerosi Multipla, Sapienza University.* Sirio Cocozza—UNRELATED: Board Membership: Amicus Therapeutics, Comments: fees for Advisory Board; Payment for Lectures Including Service on Speakers Bureaus: Sanofi, Amicus, Comments: fees for speaking. *Money paid to the institution.
Indicates open access to non-subscribers at www.ajnr.org
References
- Received November 7, 2020.
- Accepted after revision July 12, 2021.
- © 2021 by American Journal of Neuroradiology