Abstract
SUMMARY: Exponential technologic advancements in imaging, high-performance computing, and artificial intelligence, in addition to increasing access to vast amounts of diverse data, have revolutionized the role of imaging in medicine. Radiomics is defined as a high-throughput feature-extraction method that unlocks microscale quantitative data hidden within standard-of-care medical imaging. Radiogenomics is defined as the linkage between imaging and genomics information. Multiple radiomics and radiogenomics studies performed on conventional and advanced neuro-oncology image modalities show that they have the potential to differentiate pseudoprogression from true progression, classify tumor subgroups, and predict recurrence, survival, and mutation status with high accuracy. In this article, we outline the technical steps involved in radiomics and radiogenomics analyses with the use of artificial intelligence methods and review current applications in adult and pediatric neuro-oncology.
ABBREVIATIONS:
- AI
- artificial intelligence
- AUC
- area under the curve
- DL
- deep learning
- GBM
- glioblastoma
- ML
- machine learning
- SUV
- standardized uptake value
In parallel to the growing understanding of the impact of genomics and epigenomics, marked advances in clinical imaging and allied computational technologies have occurred during the past few years. These have facilitated enhanced diagnosis, molecular stratification, and targeted therapeutic clinical trials. Radiomics and radiogenomics are rapidly growing fields in imaging and, since their early inception, have been explored in the field of neuro-oncology.1 Radiomics is a high-throughput computational process that unlocks microscale quantitative data hidden within conventional imaging, not otherwise visualized by the naked human eye; radiogenomics is the linkage between imaging and genomics data.2,3 With use of radiomics analysis, a patient's scans are converted into mineable quantitative data to which machine learning (ML) techniques can be applied for integrative analysis. This process has enabled the identification of quantitative imaging markers and signature models that are reflective of microscopic tumor biology, which has led to enhanced, biologically relevant classification, tumor grading, survival prediction, and treatment response in adult and pediatric brain tumors.4⇓⇓⇓-8 Although studies of radiomics and radiogenomics have been performed in adult brain tumors, their application and use in pediatric neuro-oncology are evolving.5,9,10 Initial studies showed promising ability to distinguish posterior fossa tumors like ependymoma and pilocytic astrocytoma.9 Recent studies using a combination of radiomics and ML were able to distinguish pathologic subtypes of pediatric brain tumors.10 This article reviews the basics of radiomics, radiogenomics, and their clinical application; their evolving role in adult neuro-oncology; and their recent translation and application in pediatric neuro-oncology.
Search Criteria
Details of this review article were based on a PubMed search of categories of radiomics, adult brain tumors, and pediatric neuro-oncology. The search was limited to malignant brain tumors (excluding neurologic and benign lesions of the brain).
Technologic Methods and Computational Process
Overview of Radiomics Workflow.
The radiomics processes can be structured into multiple phases, and they are the same in pediatric and adult patients: 1) image acquisition and data selection, 2) image registration and segmentation, 3) image preprocessing, 4) radiomics feature extraction, and 5) feature selection and modeling to predict the outcomes of interest (Fig 1).
Radiomics pipeline for brain tumors. Upper line: Segmentation of the 3 imaging phenotypes: necrosis (left), enhancing tumor (middle), and edema/invasion (right). Middle and lower lines: Radiomics feature extraction, normalization, and volume-dependent feature generation are followed by feature selection and predictive modeling for outcomes.
Image Acquisition and Data Selection.
Image acquisition and data selection are considered the initial steps in the workflow. For radiomics and radiogenomics studies in neuro-oncology, MR imaging and PET are generally used.
Image Registration and Segmentation.
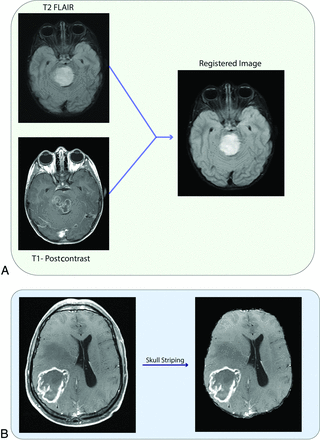
Image registration refers to the spatial alignment of the various imaging sequences into the same geometric/anatomic space (Fig 2A).11 Tumor segmentation refers to contour delineation of the volumes of interest (3D) or ROIs (2D) (Fig 3). Manual segmentation or semiautomated and automated segmentation with superimposed manual segmentation is the criterion standard. Complete manual segmentation is tedious and time-consuming and requires contouring of the tumor section by section; furthermore, it is prone to interobserver and intraobserver variability.12 Semiautomated and automated segmentation methods have been explored in various studies to reduce manual labor and improve consistency and reproducibility to overcome these challenges.12⇓-14 Although promising, semiautomated and automated methods are not entirely accurate and typically require editing by a radiologist to ensure more precise borders.13
A, Registration of T1-postcontrast MR imaging and FLAIR MR imaging. B, Skull stripping.
Segmentation and 3D volume extraction of a diffuse intrinsic pontine glioma.
Image Preprocessing.
Skull stripping: Because high-resolution structural images include a significant amount of nonbrain tissue such as bone, skin, and eyeballs, these can affect the radiomics analysis (Fig 2B). The FSL Brain Extraction Tool (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/BET) and Robust Brain Extraction (https://www.nitrc.org/projects/robex) are the commonly used algorithms for skull stripping.15 Intensity normalization: During MR imaging acquisition, the use of different scanner types and scanning parameters can result in large intensity variations. These variations can weaken the performance of subsequent radiomics analysis. It is an essential process of mapping intensities to a standard reference scale in MR imaging to account for variations between patients and longitudinal studies and to increase radiomics reproducibility. The algorithm generated by Nyúl and Udupa,16 which involves the matching of histograms, is the most commonly used normalization method. Noise reduction: Noise suggests that the pixels in the image show distinct intensity values instead of actual pixel values that are achieved from the image.17 Rician noise decreases the quality of MR imaging and makes quantitative feature extraction difficult.17 Gaussian convolution and neighborhood filter are the most commonly used noise-reduction algorithms.17 Bias field correction: The bias field signal is nonuniform low-frequency intensities that corrupt MR imaging.18 The bias field is a potential confounder for radiomics analysis, and it should be corrected. The N4 bias field correction algorithm is the most commonly used approach for inhomogeneity correction.18
Radiomics Feature Extraction.
After segmentation and preprocessing, extraction of radiomics features is then performed. Radiomics features can be categorized as first-order, second-order, and high-order features. First-order features describe the distribution of individual voxel values without concern for spatial associations between neighboring voxels.17 Second-order features calculate the statistical interrelationships between adjacent voxels. Second-order features give a measure of the spatial alignment of the voxel intensities and intralesion heterogeneity. There are multiple different ways of quantifying spatial association among pixels: Gray-level co-occurrence matrix is a most commonly used second-order statistical texture-analysis technique, which analyzes the spatial relationship among pixels and defines how frequently voxel pairs are present in different directions.17 Gray-level run length matrix, another commonly used second-order feature, quantifies consequent voxels that have the same gray-level value along a fixed direction.17 High-order features involve performing filters or mathematic transforms to the images before the feature-extraction process.19 These features aim to detect repetitive or nonrepetitive patterns, remove noise, or highlight details. The wavelet transform–based method is a very popular approach in which the image is decomposed in multiple scales and used for texture analysis.19 Laplacian transforms of the Gaussian filter identify areas of rapid intensity change (edges) in images.20,21
Artificial Intelligence Applied to Imaging: Feature Selection and Statistical Modeling
Machine Learning.
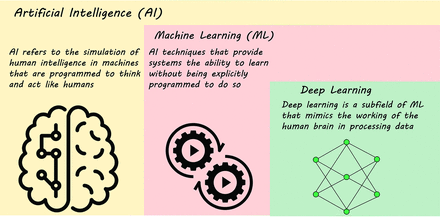
ML is a branch of artificial intelligence (AI) in which computers are given the capability of learning like humans by feeding data and information without being explicitly programmed (Fig 4).22 Fundamental to ML is that classification, regression, prediction, clustering, and association models are provided. ML methods are categorized into supervised, unsupervised, and semi-supervised learning.
AI, ML, and DL.
Supervised Machine Learning.
In supervised learning, a model is able to predict target clinical outcomes with the help of a labeled clinical dataset.22 The supervised learning model includes 2 steps: training and testing. Model training consists of inputs paired with the corresponding outputs to train the model. During training, the algorithms search for patterns in the data that relate to outcomes. Model testing is the process of predictive performance evaluation of the trained model on the test dataset. Support vector machine,8 random forests,8 and eXtreme Gradient Boosting (XGBoost; https://xgboost.ai/)23,24 are the most commonly used supervised machine learning methods in radiomics.
Unsupervised Machine Learning.
Unsupervised learning methods can classify radiomic features into subgroups using clustering algorithms such as k-means clustering,25 fuzzy clustering,26 or consensus clustering.27 Next, the relationships of the various features within their groups are compared. Subsequently, the capability of various subgroups to predict clinical output can be compared; however, labeled data are not used for the initial model development. Radiomics features can also be used to provide new subclasses that may more closely align with the underlying biology of CNS tumors by using unsupervised machine learning methods.
Deep Learning.
Deep learning (DL), also known as deep neural network, is a subfield of machine learning in AI that has multiple neural-like networks (Fig 5).28 The artificial neural networks are capable of learning from data that are unstructured or unlabeled. The most significant difference between DL and conventional ML algorithms is that DL does not need any human interference such as radiomics, feature extraction, and segmentation to learn the connection between the input and the corresponding output. Some of the known deep learning algorithms are the recurrent neural network, restricted Boltzmann machine, and convolutional neural network.28 The convolutional neural network is most commonly used and is popular in radiomics studies.28
A deep learning–based model for predicting outcomes.
Feature Selection.
Feature selection is the process of selecting a subset of the most relevant and significant discriminatory features associated with the specific outcome. These are then used for building radiomics models. Feature selection helps in clarifying the data by revealing, in an individual, the important features and how they are related to each other and to the image itself. There are several commonly used methods for radiomics feature selection such as Least Absolute Shrinkage and Selection Operator,8 Minimum Redundancy Maximum Relevance,24,29 and Generalized Linear Models with Elastic Net Penalties.30
Modeling.
Radiomics uses prediction models for predicting outcomes, including clinical or demographic, genomic, survival, response, and resistance models.31⇓⇓⇓-35 AI, specifically ML and DL, provides several approaches to achieve this aim.31 The support vector machine uses a hyperplane, which distinctly classifies the data points into 2 classes.31 Generalizability and the possibility of achieving probabilistic outputs are its advantages.31 XGBoost is a decision tree–based ensemble algorithm that uses a gradient boosting.31,36 It enables cross-validation, regularization, missing-value imputation, and flexibility.36 The choice of modeling technique has been demonstrated to affect prediction performance in a radiomics study.29
Validation.
The most effective approach for validation is the use of prospective independent external validation. In cases for which no such prospective or independent external data are available, internal validation techniques can be performed. K-fold, leave-one-out cross-validation, and hold-out are the most commonly used internal validation approaches.37
Clinical Applications of Radiomics in Neuro-Oncology: Adults
Most research in radiomics was initially performed in lung cancer.38 Radiogenomics, however, was initially performed in adult brain cancer, specifically glioblastoma (GBM).39 Since then, numerous studies have focused on the use of radiomics and radiogenomics in various adult CNS tumors (Online Supplemental Data).4,7,40⇓⇓⇓⇓⇓⇓-47 With increasing computing speed and the availability of computerized algorithms, large numbers of radiomics features can be extracted rapidly from MRIs and used to generate specific signature models. These have aided in noninvasively identifying histologic and molecular profiles of tumors,48,49 predicting response,35 differentiating pseudoprogression from true progressive disease,24 and delineating oncogenic markers in the microenvironment of brain tumors.50
Radiomics as a Diagnostic Marker.
The most recent 2016 World Health Organization classification demonstrated the importance of integrating phenotype-genotype characteristics of CNS tumors, enabling newer subclassifications of tumor groups.51 This landmark article led to seminal studies44,52,53 that showed that brain tumors could be directly related to a specific set of genomics, helping to preoperatively predict tumor genotype. GBM is the most common malignant brain tumor in adults, with poor survival outcome and a median survival of 15 months.54 Discernment of the intratumoral genetic heterogeneity of GBMs has important implications in optimizing targeted therapy to improve survival and in understanding the mechanisms of therapeutic resistance.54,55 GBMs have multiple genetically distinct clonal populations driving oncogenesis and thus have distinct therapeutic sensitivities and habitats of resistance.54 Biopsy and surgery cannot capture such spatially extensive tumor heterogeneity because both comprise samples that represent a small and static molecular “snapshot” of the entire tumor, which is very heterogeneous; furthermore, these procedures are invasive.
Radiomics and radiogenomics have been shown to potentially complement biopsy by capturing regional genetic heterogeneity and by noninvasively evaluating various driver genes and prognostic markers at diagnosis.43,44,47,56⇓-58 Recent studies have shown that radiomics models can preoperatively predict O6-methylguanine-DNA methyltransferase (MGMT) methylation, epidermal growth factor (EGFR) amplification, and EGFR variant III status in GBM.48,58⇓-60 Zhang et al46 demonstrated that ML algorithms generated from preoperative MR imaging and clinical features of 120 patients with grade III and IV gliomas predicted isocitrate dehydrogenase (IDH) 1/2 status with accuracies of 86% and 89% in the training and validation cohorts, respectively. Similarly, Chang et al61 showed the feasibility of a DL, convolutional neural network approach for the classification of genetic mutations of both low- and high-grade gliomas. They used 259 patients with either low- or high-grade gliomas to classify IDH1 mutation, 1p/19q codeletion, and MGMT methylation status. Their classification demonstrated that the convolutional neural network is capable of learning significant imaging components without prior feature selection or human-directed training with high accuracy: IDH1 mutation status, 94%; 1p/19q codeletion, 92%; and MGMT methylation status, 83%.
In a recent study, Hu et al50 built a radiomics model to preoperatively identify 6 important driver genes (EGFR, PDGFRA, PTEN, CDKN2A, RB1, and TP53) in primary GBM. The authors evaluated 48 biopsies from the regions of enhancing and nonenhancing parenchyma of 13 patients with GBM and identified significant imaging correlations for these 6 genes. They achieved accuracies ranging from 68% to 87.5% for PDGFRA, EGFR, CDKN2A, PTEN, and RB1 prediction models, whereas the accuracy for the TP53 prediction model was 37%. Similarly, Zinn et al62 showed that there are distinct MR imaging radiomics features associated with the TP53-PTEN-EGFR mutational landscape and that radiomics is approaching the complexity of whole-genome microarray expression data. Additionally, recent studies demonstrated that MR imaging–based radiomics models predicted 1p/19q codeletion status in histopathologically diagnosed gliomas.43,47 The latter is a predictor of better prognosis and a durable response to therapy.63 These results provide proof of concept and reaffirm that genomics-based biomarkers can be correlated and/or predicted noninvasively by radiomics, facilitating the evaluation of intratumoral genetic heterogeneity; and if prospectively validated, can be translated to the clinic as a noninvasive, cost-effective genomic test approach and advance individualized patient management.
Tumor Type and Tumor Grading Prediction.
Noninvasive diagnosis of brain tumors is clinically challenging but necessary because various treatment strategies are needed depending on the tumor type. Conventional or advanced MR imaging techniques are often challenging in their ability to differentiate GBM from metastatic brain tumors and lymphoma. All these tumors can show enhancement on T1-weighted images with gadolinium and hyperintensity on FLAIR.64,65 Although advanced MR imaging (MR spectroscopy and PWI) has shown promising results in the evaluation of brain tumors, further work is needed for a transition to the clinic as part of routine brain tumor management.64,65 Radiomics-based ML techniques have demonstrated potential in differentiating GBM from metastatic brain tumors.66 Another study showed that a radiomics model generated by features obtained from diffusion MR imaging yielded a better diagnostic performance than a radiomics model created by features acquired from conventional MR imaging in differentiating atypical primary CNS lymphoma, which often mimics GBM.67 Similarly, Kong et al68 showed that radiomics features obtained from the PET images have the potential to distinguish primary CNS lymphoma from GBM. They generated 3 groups of maps: a standardized uptake value (SUV) map, an SUV map calibrated with the normal contralateral cortex activity, and an SUV map calibrated with the normal brain mean activity; a total of 107 radiomics features were extracted from each SUV map. Their areas under the curve (AUCs) for differentiating CNS lymphoma and GBM ranged from 0.644 to 0.999. In addition, a recently published study determined the capability of radiomics analysis to identify the primary origin of brain metastases despite neuropathologic procedures and imaging evaluations often failing to identify the primary tumor site and leading to delayed diagnosis and treatment.69 However, further studies are needed to validate and reproduce these findings.
High-grade gliomas, including GBMs, are biologically elusive, due to their genetic heterogeneity and complex imaging phenotypes.54,55 Previous studies have shown that radiomics can predict the pathologic behavior of gliomas, histologic/molecular subtypes/grading, and the proliferative index.49,70,71 This finding could facilitate the presurgical evaluation of tumor behavior, determine the extent of surgical resection, and enable optimal clinical treatment decision-making in patients with various grades of gliomas. Histologic grade and the Ki-67 labeling index are used to help predict biologic behavior and prognosis of gliomas.72 A recently published study has demonstrated that radiomics features based on T2 FLAIR images have been shown to preoperatively correlate and predict multiple immunohistochemical features of gliomas such as Ki-67, S-100, vimentin, and CD34 expression.49 These insights are expected to enhance the personalized treatment of patients with gliomas.49 Furthermore, radiomics-based ML techniques have shown potential in differentiating GBM from anaplastic oligodendroglioma.42 Although the AUCs of the models used were >0.900, more studies in a larger cohort of patients are required to further validate these revelations.
Risk Stratification and Prognostication of Tumors.
The 2016 World Health Organization classification considers the genomics heterogeneity of diffuse gliomas, necessitating effective risk stratification for optimizing therapy.51 The invasiveness of existing methods to identify histopathologic/molecular profiles and the difficulty of replicating results demonstrated the need to identify noninvasive methods to complement and solidify the diagnostic armamentarium. Development of radiomics profiles and comprehensive analysis of radiogenomics phenotypes using multiomics molecular approaches have been successful in risk stratification and survival estimation of diffuse gliomas.6,32,33,35,41,73,74 A study using MR imaging radiomics features of patients with de novo GBM determined the imaging signatures associated with poor prognosis.75
In another cohort of 82 patients with de novo GBM, analysis of 5 texture features from preoperative MR images predicted molecular subtypes and 12-month survival status (overall survival at 12 months indicating whether the patient was alive).32 Another radiomics study evaluating CBV and ADC in 119 patients with newly diagnosed GBM demonstrated that radiomics-based classification allows noninvasive prediction of survival and stratification of patients with GBM with better accuracy than that determined with established conventional clinical (age and Karnofsky Performance Scale score) or radiologic risk models (Gaussian normalized relative CBV and ADC).6 These methods and tools could guide planning of surgical resection, define radiation treatment margins, and optimize the intensity of chemotherapy. The evolving and molecular profile of lower-grade gliomas, including the favorable outcome associated with IDH1 and 1p/19q codeletion mutation status, now partially explains the heterogeneous survival outcomes; recent studies have shown that radiomics phenotyping using ML techniques can identify these genomic markers43,47 and predict overall survival with better accuracy than with the use of nonimaging markers (clinical features and genomic data).41 Identification of the ATRX mutation or loss of ATRX expression is a potential biomarker in gliomas and has been associated with favorable survival outcome; in a recently published article, the authors showed that radiomics could identify this genotype at diagnosis.56 This could be an important addition to the noninvasive detection of pivotal biomarkers such as IDH1 and MGMT in gliomas.44,46,47,58,59
Predicting Treatment-Related Surrogate End Points.
A few studies have robustly defined the biologic heterogeneity of the peritumoral brain zone and its interaction with the microenvironment in brain tumors.55 This heterogeneity contributes to therapeutic resistance and poor survival in infiltrating tumors such as GBM.55 Nearly 90% of recurrences in GBM occur in the peritumoral brain zone, which is usually the nonenhancing component.55 Interrogating 10 radiomics peritumoral features from the peritumoral brain zone on routine preoperative MR imaging in patients with GBM was predictive of long-term (>18 months) versus short-term (<7 months) survival.33 In addition, a model generated by 6 imaging features for CD3 infiltration prediction in GBM achieved high accuracies of 97.1% and 76.5% in the training and testing sets, respectively.76 The role of radiomics is being used increasingly to improve characterization and understanding of the tumor microenvironment.70,76 This will have important clinical relevance in optimizing a targeted therapeutic approach.77
Bevacizumab has been widely used to treat patients with recurrent GBM; however, the data show a lack of sustained long-term efficacy.78 The therapeutic responses, furthermore, vary substantially in patients.78 However, quantitative imaging biomarkers that use radiomics to help identify patients who will benefit from bevacizumab treatment before initiation of therapy have been researched.35
With the use of immunotherapeutics in brain tumors, transient increases in tumor size and/or new inflammatory lesions often appear. These changes, known as pseudoprogression, typically stabilize or decrease with the continuation of treatment, and differentiating them from progressive disease is often difficult and delayed.79 Conventional imaging using MR perfusion studies has been used frequently, though specificity and sensitivity remain suboptimal.80,81 A multicenter study using an MR perfusion–based radiomics model was able to distinguish pseudoprogression from progressive disease with the AUC, sensitivity, and specificity of 90%, 93%, and 89%, respectively.24 These studies highlight the capability of radiomics to address this important clinical distinction—an unmet need in neuroimmunotherapy.
Evolving Era of Radiomics in Pediatric Neuro-Oncology
Pediatric CNS tumors have special characteristics in tissues, morphology, molecular subtype, and texture compared with their adult counterparts.82 With the emerging biologic differences between adult and pediatric brain tumors, the need for improved characterization of CNS tumors in children is imperative. Although radiomics and radiogenomics are now used in the adult clinical arena, their translation into clinical practice in pediatric neuro-oncology is still in its infancy (Online Supplemental Data). Pediatric brain tumors are the most common solid tumors in children.83 Unlike adults, in whom we have seen increased use of noninvasive neuroimaging using multimodal MR imaging combining multiple sequences to enhance tissue characterization, few studies to date have been reported in pediatrics.5,9,84⇓⇓-87 Similar noninvasive imaging studies are needed in parallel with the evolving molecular era in pediatric brain tumors, facilitating improved tumor classification (histologic and molecular), grading, survival prediction, and treatment response.
Earlier studies evaluated the role and potential of MR imaging texture analysis in capturing quantitative information of different pediatric brain tumors.88 A recent multicenter study was performed within a supervised classification framework on clinical MR imaging, and a support vector machine was trained with 3D textural attributes obtained from conventional MR imaging.85 The developed model was very successful in capturing transferable tumor information, and this study supported the use of 3D texture analysis on conventional MR imaging to aid in the diagnostic classification of various pediatric brain tumors. A recently published study84 using a combination of radiomics and ML approaches on 3D multimodal MR imaging was able to build a radiomics model with multivariable logistic regression. These models could differentiate pediatric ependymoma and medulloblastoma (common malignant brain tumors of the posterior fossa in children) with an AUC of 0.91. Similarly, Zhou et al87 demonstrated the capability of radiomics in differentiating the types of pediatric posterior fossa tumors (medulloblastoma, ependymoma, and pilocytic astrocytoma) with an accuracy of 0.85. The success of this trend needs to be evaluated further in prospective studies.
Pediatric medulloblastoma has been the most biologically interrogated malignant CNS tumor in children.89 Its molecular subtypes with distinct clinical and prognostic differences now reaffirm the need to tailor therapy accordingly.89 A recent study5 provided proof-of-concept results for the application of radiomics and ML approaches for prediction of distinct medulloblastoma subgroups. The reproducibility of this finding would be of immense clinical significance, with noninvasive ability to diagnose and biologically subtype these neoplasms. Another research group published that model based on texture extracted from conventional MR imaging to preoperatively differentiate ependymoma and pilocytic astrocytoma and achieved high sensitivity, specificity, and AUC.9 This could be an invaluable tool because often these lesions occur in the eloquent areas of the brain both at diagnosis and recurrence, precluding surgical intervention.
DNA methylation now identifies various molecular subtypes of aggressive pediatric CNS tumors, and genomics analysis was able to decipher the intrinsic tumor heterogeneity.90 Noninvasive imaging that uses radiomics to correlate with these genomic aberrations would be a clinically significant addition to diagnosing these neoplasms and facilitating the design of optimal targeted therapy.
Diffuse midline gliomas in children are aggressive brain tumors with 10% overall survival at 18 months. MR imaging–based texture analysis in 32 children with diffuse midline gliomas was able to stratify patients into poor and good prognostic groups, with a median survival of 7.5 months versus 17.5 months, respectively.74 Diffuse midline gliomas with more homogeneous texture on diagnostic MR imaging were associated with a worse prognosis.74 These findings should be further explored and correlated with the evolving molecular subtypes, providing valuable insight into prognosis and tailored therapy for these tumors. Profiling an optimal decision tree model with the use of ADC histogram analysis and structural MR imaging findings provided the ability to differentiate 7 histopathologic subtypes of pediatric CNS tumors, with a nearly 90% accurate classification.34 This decision tree model could preoperatively distinguish, with precision, 2 biologically different tumors, pilocytic astrocytomas and atypical teratoid/rhabdoid tumor, in comparison with neuroradiologists. Similar prospective studies evaluating the ability of this model to correlate with the diverse genomic aberrations would be a breakthrough in the management of targeted therapy for these tumors at diagnosis and recurrence. A major success of targeted therapy in pediatric CNS tumors has been the use of BRAF and Mek inhibitors in low-grade gliomas. Currently, a biopsy is needed to identify these molecular aberrations, often not possible due to the eloquent location of these neoplasms. A recent study showed the ability of radiomics-based prediction of BRAF status in pediatric low-grade gliomas before a biopsy, which could have significant implications in clinical practice.91
Immunology.
Use of immunotherapeutic strategies, including adoptive cell therapy, vaccine therapy, and checkpoint inhibitors, in pediatric brain tumors has been rapidly evolving.92 However, therapeutic efficacy is undermined by the low mutational burden seen in these tumors, precluding targeted therapy. Reviews of immunotherapy in pediatric brain tumors show some preliminary promising results, which need to be further explored. In a recently published study of natural killer cell infusion in children with recurrent medulloblastoma and ependymoma, a high radiomic performance was achieved; accuracy, sensitivity, and specificity were 100%, though it did not attain statistical significance, likely due to the low number of patients.86,92,93 A few studies in adults have shown the ability of radiomics models to differentiate pseudoprogression from early tumor progression in infiltrative tumors such as GBM.24 The ability to make these distinctions in pediatric neuro-oncology is important and needs to be pursued.
Limitations
Although radiomics has shown its potential for diagnostic, prognostic, and predictive purposes in multiple neuro-oncology studies, the translation of radiomics into the clinical settings is slow because the field is facing several challenges. Most important, the repeatability, variability, and reproducibility of radiomics are still issues and often depend on the imaging sequence used, technique, size of the image, image quality, software used for feature extraction and reconstruction, as well as motion artifacts and segmentation, which can lead to greater interobserver variations in addition to the time-consuming nature of manual segmentation.
DL-based radiomics may overcome these issues because the features are extracted automatically from the huge data, and a subset of best-performing features is automatically identified. The most important pitfall of DL-based radiomics is the high association between the images and the clinical output data. Thus, in contrast to feature-based radiomics, large datasets are needed to identify the most relevant and robust features to overcome this challenge. DL-based radiomics models must validate their reliability in prospective large studies.
Another important limitation to the translation of radiomics models in clinical management is the interpretability of the radiomics features and models. Radiomics is perceived as a “black box,” ie, it is very difficult to clinically interpret the developed predictions. As an emerging technique, explainable AI is better understood, trusted, and the results are more efficiently interpreted by human-users, in contrast to the “black box” concept in ML.94
Conclusions
In this review, we outlined the principles of radiomics analysis in a step-by-step approach and described how large amounts of imaging data can be interrogated and analyzed in patients with brain tumors. We believe that soon, with cumulative developments in AI and increasing public access to large data bases, radiomics can be expected to play an important role in clinical decision-making by answering many clinical questions of diagnostic and therapeutic relevance. However, many limitations and challenges exist, and much work needs to be done to robustly translate radiomics into the clinical arena.
Footnotes
Rivka R. Colen and Soumen Khatua are senior co-authors.
This work was funded, in part, by startup funding from The University of Pittsburgh Hillman Cancer Center (R.R.C.) and the Hillman Cancer Center's National Cancer Institute Cancer Center Support Grant No. P30CA047904 (R.R.C.).
Indicates open access to non-subscribers at www.ajnr.org
References
- 1.↵
- 2.↵
- 3.↵
- 4.↵
- 5.↵
- 6.↵
- 7.↵
- 8.↵
- 9.↵
- 10.↵
- 11.↵
- 12.↵
- 13.↵
- 14.↵
- 15.↵
- 16.↵
- 17.↵
- 18.↵
- 19.↵
- 20.↵
- 21.↵
- 22.↵
- 23.↵
- 24.↵
- 25.↵
- 26.↵
- 27.↵
- 28.↵
- 29.↵
- 30.↵
- 31.↵
- 32.↵
- 33.↵
- 34.↵
- 35.↵
- 36.↵
- 37.↵
- 38.↵
- 39.↵
- 40.↵
- 41.↵
- 42.↵
- 43.↵
- 44.↵
- 45.↵
- 46.↵
- 47.↵
- 48.↵
- 49.↵
- 50.↵
- 51.↵
- 52.↵
- 53.↵
- 54.↵
- 55.↵
- 56.↵
- 57.↵
- 58.↵
- 59.↵
- 60.↵
- 61.↵
- 62.↵
- 63.↵
- 64.↵
- 65.↵
- 66.↵
- 67.↵
- 68.↵
- 69.↵
- 70.↵
- 71.↵
- 72.↵
- 73.↵
- 74.↵
- 75.↵
- 76.↵
- 77.↵
- 78.↵
- 79.↵
- 80.↵
- 81.↵
- 82.↵
- 83.↵
- 84.↵
- 85.↵
- 86.↵
- 87.↵
- 88.↵
- 89.↵
- 90.↵
- 91.↵
- 92.↵
- 93.↵
- 94.↵
- Received February 1, 2021.
- Accepted after revision July 19, 2021.
- © 2022 by American Journal of Neuroradiology