Abstract
BACKGROUND AND PURPOSE: The current imaging assessment of fetal brain gyrification is performed qualitatively and subjectively using sonography and MR imaging. A few previous studies have suggested methods for quantification of fetal gyrification based on 3D reconstructed MR imaging, which requires unique data and is time-consuming. In this study, we aimed to develop an automatic pipeline for gyrification assessment based on routinely acquired fetal 2D MR imaging data, to quantify normal changes with gestation, and to measure differences in fetuses with lissencephaly and polymicrogyria compared with controls.
MATERIALS AND METHODS: We included coronal T2-weighted MR imaging data of 162 fetuses retrospectively collected from 2 clinical sites: 134 controls, 12 with lissencephaly, 13 with polymicrogyria, and 3 with suspected lissencephaly based on sonography, yet with normal MR imaging diagnoses. Following brain segmentation, 5 gyrification parameters were calculated separately for each hemisphere on the basis of the area and ratio between the contours of the cerebrum and its convex hull. Seven machine learning classifiers were evaluated to differentiate control fetuses and fetuses with lissencephaly or polymicrogyria.
RESULTS: In control fetuses, all parameters changed significantly with gestational age (P < .05). Compared with controls, fetuses with lissencephaly showed significant reductions in all gyrification parameters (P ≤ .02). Similarly, significant reductions were detected for fetuses with polymicrogyria in several parameters (P ≤ .001). The 3 suspected fetuses showed normal gyrification values, supporting the MR imaging diagnosis. An XGBoost-linear algorithm achieved the best results for classification between fetuses with lissencephaly and control fetuses (n = 32), with an area under the curve of 0.90 and a recall of 0.83. Similarly, a random forest classifier showed the best performance for classification of fetuses with polymicrogyria and control fetuses (n = 33), with an area under the curve of 0.84 and a recall of 0.62.
CONCLUSIONS: This study presents a pipeline for automatic quantification of fetal brain gyrification and provides normal developmental curves from a large cohort. Our method significantly differentiated fetuses with lissencephaly and polymicrogyria, demonstrating lower gyrification values. The method can aid radiologic assessment, highlight fetuses at risk, and may improve early identification of fetuses with cortical malformations.
ABBREVIATIONS:
- AUC
- area under the curve
- GA
- gestational age
- GI
- gyrification index
- GIA
- gyrification index by area
- GIC
- gyrification index by contour
- LIS
- lissencephaly
- MCC
- Matthews correlation coefficient
- max
- maximum
- MCD
- malformation of cortical development
- PMG
- polymicrogyria
- SI
- symmetry index
Fetal cortical development is a complex process that relies on adequate cell differentiation, proliferation, neuronal migration, and organization.1 Gyrification starts gradually from the second trimester and continues during postnatal life.2 The spatiotemporal pattern of normal fetal gyrification is correlated with gestational age (GA) and is used to evaluate brain development.3⇓⇓-6
Disruption of this organized process can cause malformations of cortical development (MCDs). Two common MCDs are lissencephaly (LIS) and polymicrogyria (PMG). In LIS, the brain appears smooth with complete or partial gyral loss,6 while PMG is characterized by excessive, irregular small gyri that can be focal, multifocal, unilateral, or bilateral.7 MCDs are associated with a wide range of clinical outcomes and neurodevelopment disorders later in life.8
Fetal development is assessed primarily by using sonography, with MR imaging as a complementary tool to support its findings or when clinically indicated,9⇓⇓⇓-13 and was shown to have significant advantages in diagnosing brain malformations.14⇓-16 Clinical assessment of fetal brain gyrification using both sonography and MR imaging is qualitative through visual inspection of the cortical global appearance and specific sulci such as the Sylvian fissure, and it requires special expertise and experience.7,17,18
The gyrification index (GI) was first introduced in histologic images of adult brains,17 defined as the ratio between the outer cortical contour and the cerebral hull contour tightly wrapping the brain. Additional quantitative parameters such as curvature, surface area, and the 3D GI have been proposed for the 3D-reconstructed cortical surface.18⇓⇓-21 Global and local 3D GIs were found to correlate with age (in children),22 sex,23 and cognitive functions (in adults).24 Furthermore, parameters from other research fields have been applied to adults.4,19,25⇓⇓-28 However, these methods are less applicable to fetal brain MR imaging, due to changes in GA; high variability in scanning planes, resolutions, and contrasts, as well as changes in fetal position and motion artifacts.
Some studies proposed methods to quantify fetal gyrification on the basis of the 3D MR imaging–reconstructed cortical surface.20,28⇓⇓⇓-32 However, these methods require unique data acquisition, with a few volumes acquired in different spatial planes, and they are time-consuming and sensitive to fetal motion and require high computational power. In addition, these methods were developed in small cohorts (<40, except as in Wright et al20) based on specific MR imaging sequences with only a few proposed developmental curves for a narrow GA range (except in Wright et al20).
Our aims were the following: 1) to develop an automatic pipeline for quantitatively assessing fetal cortical folding on the basis of clinical 2D MR imaging data; 2) to quantify normal gyrification and symmetry on a large cohort of control fetuses; and 3) to measure gyrification in fetuses with LIS and PMG and evaluate the machine learning classification models for control fetuses and fetuses with LIS or PMG based on quantitative gyrification parameters.
MATERIALS AND METHODS
This retrospective study was approved by the local ethics committees of 2 institutions, which waived the need for informed consent.
MR Imaging Data and Study Population
The data set included coronal T2-weighted MR imaging, acquired between 2007 and 2022 from 2 clinical sites, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel, and the Children's Hospital of Eastern Ontario, Ottawa, Ontario, Canada, using 6 MR imaging scanners from 2 vendors and different sequences and parameters, as shown in Table 1.
MR imaging data included in this study
Control fetuses were chosen according to GA for a wide and well-represented range of weeks with normal MR imaging findings, no chromosomal abnormalities, and no evidence of fetal cytomegalovirus infection or chronic maternal disease. Pregnancies were dated according to the first-trimester crown-rump length. Clinical indications for MR imaging in controls included a suspected fetal abnormality on sonography, maternal cytomegalovirus seroconversion, a family history of genetic disease, and previous pregnancy with confirmed abnormalities. Fetuses with MCD were diagnosed by an expert fetal MR imaging neuroradiologist (L.B.-S. with > 20 years' experience or E.M. with >15 years).
Poor-quality images, including severe motion or artifacts, were excluded.
Image Analysis
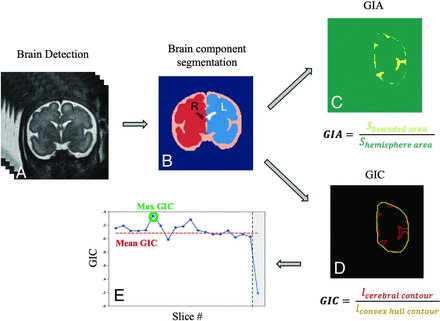
The image-processing pipeline (Fig 1) includes 4 stages:
A) Brain detection as previously described in Dudovitch et al33 using an anisotropic 3D U-Net classifier for initial segmentation and computing a tight bounding box around it.
B) Brain component segmentation,34 using a 2D multiclass segmentation network, dividing the fetal brain into 6 components: left/right hemispheres, left/right lateral ventricles, extra-axial cerebrum-spinal fluid, and cerebellum (including the brainstem). Anatomic left and right classifications were based on the inferior-superior direction (cerebellum) and anterior-posterior direction (eyes, automatically identified using another deep learning network). Manual segmentation corrections were performed when needed to ensure accurate GI calculation.
Image-analysis pipeline: A, Brain detection. B, Brain component segmentation. C, GIA calculation based on the bounded area (yellow) between the cerebral cortex and its convex hull and the hemisphere area (green, surrounded by the yellow area). D, GIC calculation based on contour extraction of the cerebral hemisphere (red) and its convex hull (yellow). E, Example of GIC values in all slices of a single fetus. Outliers are marked in gray and were not included in the mean GIC.
Next, the cerebral contour was defined as the boundary between the cerebrum and the extra-axial cerebrum-spinal fluid, and the convex-hull was computed as the smallest convex that contains the contour of the hemisphere. GI parameters were calculated for each slice, separately for the right and left side as follows:
C) GI by area (GIA) was defined as the ratio of the bounded area between the cerebral cortex and convex hull contours and the hemisphere area.
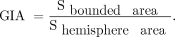 Equation 1
Equation 1
D) GI by contour (GIC) was defined as the ratio of the cerebral and convex hull contour lengths.
 Equation 2
Equation 2
We extracted 5 gyrification parameters: 1) mean GIA, the mean value from all slices excluding outliers. A predefined threshold was used and slices located at the most anterior or posterior parts with the GI below this threshold were excluded; 2) maximum (max) GIA, the maximum value from all slices, hypothesized to represent the Sylvian fissure level; 3) bounded volume, the bounded volume between the hemisphere and its convex hull, normalized by the hemisphere volume. Similarly, 4) mean GIC and 5) max GIC were extracted on the basis of the GIC defined above.
In addition, the symmetry index (SI) was calculated for each parameter, where R and L are the GIs of the right and left hemispheres, respectively:
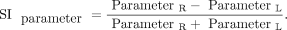 Equation 3
Equation 3
Statistical Analysis
Statistical analysis was performed using R Studio, Version 022.07.2 (http://rstudio.org/download/desktop).
Changes with GA in the control group were assessed by fitting a second-degree polynomial using a parametrical Generalized Additive Models for Location, Scale, and Shape (GAMLSS; https://www.gamlss.com/) with a Box-Cox power exponential distribution, as recommended by the World Health Organization.35 All P values were adjusted for multiple comparisons as described by Benjamini and Yekutieli.36 The 95% nonparametric confidence intervals37 were adjusted to a false coverage rate of 0.05.38
MCD Classification
Differences between control fetuses and MCDs were first assessed using Wilcoxon rank-sum tests while controlling for GA. Next, a machine learning classifier was developed to differentiate between fetuses with normal gyrification and those with MCDs, separately for LIS and PMG, based on all the gyrification parameters presented in this study: right and left GIA max, GIA mean, GIC max, GIC mean, and bounded volume. The control group included fetuses older than 24 weeks' GA (n = 86) to match the MCD group, because MCDs can only be diagnosed after 24 weeks' GA.39,40 Seven models were tested, including random forests41 (n = 100), linear and radial basis function kernel Support Vector Machine (SVM),42 Extreme Gradient Boosting (XGBoost; https://www.nvidia.com/en-us/glossary/data-science/xgboost/)43 using 2 different objectives, logistic and linear k-nearest neighbors (k = 5), and multilayer perceptron. To compensate for the inherent imbalance between control and pathologic cases, the Synthetic Minority Oversampling Technique (SMOTE; https://arxiv.org/abs/1106.1813)44 was used in model training. The training phase was performed using a 3-fold cross-validation scheme, randomly splitting the data into training data sets (LIS: n = 66, fifty-eight controls and 8 cases of LIS; PMG: n = 66, fifty-eight controls and 8 cases of PMG) and validation data sets (LIS: n = 32, twenty-eight controls and 4 cases of LIS; PMG: n = 33, twenty-eight controls and 5 cases of PMG). The performance of the models was evaluated using several metrics, including accuracy (Equation 4), F1 score (Equation 5), Matthews correlation coefficient (MCC) (Equation 6),45 area under the curve (AUC) of the receiver operating characteristic curve, precision, and recall (Equations 7 and 8):
 Equation 4
Equation 4
 Equation 5
Equation 5
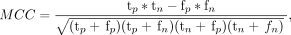 Equation 6
Equation 6
 Equation 7
Equation 7
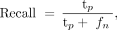 Equation 8 where tp is true-positive, tn is true-negative, fp is false-positive, and fn is false-negative.
Equation 8 where tp is true-positive, tn is true-negative, fp is false-positive, and fn is false-negative.
RESULTS
A data set of 200 fetuses was collected, with 9 fetuses excluded due to poor image quality and 29 for genetic or abnormal MR imaging findings. Of the 162 fetuses finally included, there were 134 controls (mean GA, 27.9 [SD, 5.3] weeks; range, 18–36 weeks); 12 with LIS (mean GA, 28.3 [SD, 3.2] weeks; range, 33–24 weeks); 13 with PMG (mean GA, 31.5 [SD, 2.7] weeks; range, 27–37 weeks); and 3 fetuses suspected of having LIS on the basis of sonography (mean GA, 31.3 [SD, 1.1] weeks; range, 30–32.5 weeks), but with normal MR imaging findings.
Controls
Figure 2 shows 5 control fetuses at different GAs with right hemisphere contours at the Sylvian fissure level, demonstrating the advancement in gyrification with gestation. Developmental curves for GIA and GIC parameters are presented in Figs 3 and 4, respectively, separated by the right and left hemispheres. All GI parameters change significantly throughout gestation (P < .05).
T2-weighted MR imaging of coronal views for control fetuses demonstrating gyrification development of the right hemisphere (red, hemisphere contour) at the Sylvian fissure level with gestation.
Development curves with GA of area-based gyrification parameters. Curve percentile lines 3, 15, 50, 85, and 97 are presented as blue, light blue, purple, green, and yellow, respectively. Note control fetuses (gray), PMG (green), LIS (red), and suspected LIS (orange).
Development curves with GA of contour-based gyrification parameters. Curve percentile lines 3, 15, 50, 85, and 97 are presented as blue, light blue, purple, green, and yellow, respectively. Note control fetuses (gray), PMG (green), LIS (red), and suspected LIS (orange).
Most parameters increased throughout gestation, mainly from 23 weeks onward. The max GIA exhibited a U-shaped parabolic curve, reaching a minimum around the 25th week of GA, possibly due to changes in the Sylvian fissure.
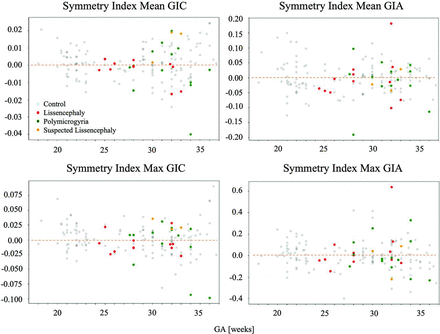
Symmetry indices with GA are shown in Fig 5, demonstrating homogeneous dispersion, indicating no brain asymmetry.
Symmetry indices for GI parameters with GA. All are close to zero, with homogeneous dispersion, indicating no brain asymmetry.
MCD
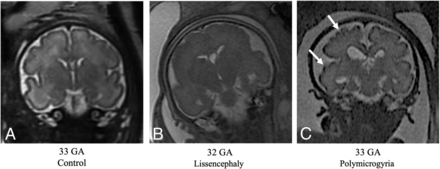
Fetuses with MCD exhibited underdeveloped gyrification compared with controls. All GI parameters were significantly lower in fetuses with LIS (P < .02) highlighted at late GA, as seen in Figs 3 and 4, and Table 2. Representative MR imaging of controls and fetuses with LIS and PMG of equivalent ages are demonstrated in Fig 6.
T2-weighted MR imaging of coronal views for controls and fetuses with LIS and PMG of equivalent ages. A, Control fetus, 33 weeks' GA. B, Fetus diagnosed with LIS at 32 weeks' GA, characterized by undeveloped gyrification patterns. C, Fetus diagnosed with PMG at 33 weeks' GA, with abnormal excessive gyri (white arrows).
Comparisons between LIS or PMG with control fetuses—the adjusted P values and CIs
Three fetuses suspected of having LIS based on sonography but diagnosed with normal MR imaging, findings showed normal GI values, supporting the MR imaging diagnosis.
Fetuses with PMG had significantly lower GICs (P < .001), with no significant difference in GIAs compared with controls (Table 2).
Brain asymmetry was not significant in fetuses with MCD, with homogeneous dispersion and no change in GA (Fig 5). However, 2 fetuses with PMG showed extreme asymmetry, confirming the radiologic diagnosis. One fetus (34 GA) had PMG only in the right hemisphere, while the other fetus (36 weeks' GA) had PMG focused anteriorly, with an abnormal shape of the right operculum.
MCD Classification
Using machine learning classifiers based on GIC and GIA parameters, we evaluated 7 models. The performance of all models is shown in the Online Supplemental Data. The best results for classification of controls and fetuses with LIS were achieved using XGBoost-logistic, with the F1 score = 0.60, accuracy = 0.87, AUC = 0.90, MCC = 0.56, precision = 0.48, and recall = 0.83. The best classifier used for PMG was random forest with an F1 score = 0.59, accuracy = 0.89, AUC = 0.84, MCC = 0.53, precision = 0.57 and recall = 0.61.
DISCUSSION
In this study, we developed a method for automatic quantification of fetal gyrification based on clinical heterogeneous 2D MR imaging data, demonstrating high robustness, and we proposed 5 parameters. We found significant changes in gyrification with GA in control fetuses, with no asymmetry. We also found significantly reduced gyrification in fetuses with MCD compared with controls.
Five quantitative gyrification parameters were extracted for each hemisphere, including the contour-based parameters GIC and area-based parameters of GIA. Previous studies used different methods, including ridge detection,28 local tensor-based morphometry,29 and curvature-based20,30 and sulcal pattern similarities with fetal brain atlas.31 However, all these methods require 3D reconstruction, are time-consuming, and are not easily applicable to clinical data. Our proposed method presents simple quantifying parameters that can be used in routine clinical practice and applied to retrospective data of large cohorts.
Developmental Curves
In this study, growth curves were calculated as recommended by the World Health Organization.35 Our results indicate that gyrification in control fetuses increases in a second-degree polynomial curve after the 23rd week of gestation. Our findings corroborate previous studies by Dubois et al46 who described cortical development in preterm infants across a similar range, demonstrating an accelerated gyrification measured by the cortical surface area and its GI after 28 weeks. In addition, Wright et al20 showed nonlinear increased values in several global curvature–based parameters. Similarly, Rajagopalan et al28 found that the GI development based on 3D reconstruction was best described with a second-order polynomial curve. Only 1 study demonstrated a linear gyrification increase with GA, probably due to the narrow range of GAs.29
Brain Symmetry
Our study did not find evidence of gyrification asymmetry, consistent with previous studies that quantified global gyrification on the basis of a 3D reconstructed cortical surface.18,20,22,23,25,26,28⇓-30,47 While there is known asymmetry in specific sulci, such as the superior temporal gyrus,3,46,48 our results suggest that global gyrification measurements may not be sensitive enough to detect subtle regional differences.
MCD
Our results demonstrate significantly reduced gyrification in fetuses with LIS, with greater deviations at advanced GA. Notably, our results showed values within the normal range in 3 fetuses suspected of having LIS on the basis of sonography, yet with a normal MR imaging diagnosis.
Reduced gyrification was also detected in fetuses with PMG, but only in the GIC parameters. One previous study31 quantified gyrification in PMG and normally developing fetuses using 3D GI measurements yet did not find differences between the 2 groups. This was a small-scale study (Ncontrol = 17, NPMG = 3). To our knowledge, no additional studies quantified LIS and PMG fetal gyrification patterns with gestation or developed classifiers on the basis of control fetuses and fetuses with MCD. The classifiers trained in this study, both for LIS and for PMG, showed good performance in several metrics; however, the F1 score was improved when using SMOTE for imbalanced data and showed low values. These results are expected due to the overlap of MCDs and healthy fetuses in some of our gyrification parameters. Our findings support the use of quantitative analysis for fetal brain assessment in fetuses with MCD, to highlight fetuses at risk and to aid radiologic interpretation and diagnosis.
Limitations
Our study limitations include the potential impact of section acquisition symmetry on 2D gyrification parameters. However, analyzing each hemisphere separately may address this issue. This study focused on global parameters and did not analyze specific sulci. Future studies should develop automatic 2D methods to assess local parameters in different regions. Additionally, our gyrification parameters may pseudonormalize the multiple and shallow gyri in PMGs, leading to values within the normal range. The outcomes of the MCD fetuses were unavailable, and our criterion standard was the radiologist's diagnosis based on MR imaging. Finally, our cohort included 25 fetuses with MCD, representing the largest study to date that quantifies gyrification in MCD, yet the number is relatively small, especially when developing an automatic classifier. Therefore, due to the small cohort size, the classifiers developed in this study were only validated and not tested on a separate test set. Moreover, higher recall is needed and other parameters should be included in the model such as brain volume, brain biometrics, and so forth, to develop a method for clinical use.
CONCLUSIONS
This study presents an automatic quantification of fetal brain gyrification based on 2D routinely acquired MR imaging data and suggests the use of 5 parameters. The method successfully detected changes in normal gyrification with GA and showed reduced gyrification in fetuses with MCD compared with controls. These findings suggest that quantifying gyrification can aid in assessing fetal brain maturation and identifying MCDs.
ACKNOWLEDGMENTS
This research was supported by the Israel Innovation Authority and the Yoran Institute of Human Genome Research. Mr Yair Wexler, School of Neurobiology, Biochemistry and Biophysics, Faculty of Life Sciences, Tel Aviv University, Tel Aviv, Israel, provided statistical advice for this study.
Footnotes
This work was supported by the Yoran Institute of Human Genome Research and the Israel Innovation Authority.
Disclosure forms provided by the authors are available with the full text and PDF of this article at www.ajnr.org.
Indicates open access to non-subscribers at www.ajnr.org
References
- Received May 3, 2023.
- Accepted after revision September 23, 2023.
- © 2023 by American Journal of Neuroradiology